Activity of the human papillomavirus type 16 late negative regulatory element is partly due to four weak consensus 5' splice sites that bind a U1 snRNP-like complex - PubMed (original) (raw)
Activity of the human papillomavirus type 16 late negative regulatory element is partly due to four weak consensus 5' splice sites that bind a U1 snRNP-like complex
Sarah A Cumming et al. J Virol. 2003 May.
Abstract
The human papillomavirus (HPV) life cycle is tightly linked to differentiation of the squamous epithelia that it infects. Capsid proteins, and hence mature virions, are produced in the outermost layer of differentiated cells. As late gene transcripts are produced in the lower layers, posttranscriptional mechanisms likely prevent capsid protein production in less differentiated cells. For HPV type 16 (HPV-16), a 79-nucleotide (nt) negative regulatory element (NRE) inhibits gene expression in basal epithelial cells. To identify key NRE sequences, we carried out transient transfection in basal epithelial cells with reporter constructs containing the HPV-16 late 3' untranslated region with deletions and mutations of the NRE. Reporter gene expression was increased over 40-fold by deletion of the entire element, 10-fold by deletion of the 5' portion of the NRE that contains four weak consensus 5' splice sites, and only 3-fold by deletion of the 3' GU-rich region. Both portions of the element appear to be necessary for full repression. Inactivating mutations in the 5' splice sites in the 5' NRE partially alleviated repression in the context of the 79-nt NRE but caused full derepression when assayed in a construct with the 3' NRE deleted. All four contribute to the inhibitory effect, though the second splice site is most inhibitory. Sm proteins, U1A and U1 snRNA, but not U1 70K, could be affinity purified with the wild-type NRE but not with the NRE containing mutations in the 5' splice sites, indicating that a U1 snRNP-like complex forms upon the element.
Figures
FIG. 1.
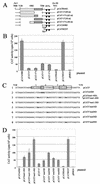
Diagram of the HPV-16 genome and sequence of the region containing the NRE and its putative secondary structure plot. (A) The linearized genomic structure of HPV-16, showing early (E) and late (L) gene coding regions (boxed). P97, constitutively active promoter; P670, promoter activated in differentiated cells; arrowheads [p(A)E and p(A)L], early and late poly(A) signals, respectively; heavy line, noncoding region (NCR). (B) The sequence of the 445-nt _Pst_I-_Eco_RI fragment in the L1-late 3′ UTR of the viral genome. Boldface, L1 stop codon; boxed region, NRE; stippled box, loop of the predicted stem-loop structure; gray box, GU-rich region; diamonds, exon-intron boundaries of weak consensus 5′ splice sites; underlined sequences, late polyadenylation signals, LP1 and LP2; overlined sequences, extent of 5′ and 3′ stems of the stem-loop structure. (C) High-energy Zuker fold model of the 445-nt _Pst_I-_Eco_RI fragment in the L1-late 3′ UTR of the viral genome. Every 50 bases are numbered. The 5′ and 3′ ends of the NRE are indicated with arrows.
FIG.2.
Functional analysis of mutant NRE constructs. (A) NRE deletion mutants. Open box, 5′ NRE; hatched box, 3′ GU-rich portion of NRE; arrowheads, late polyadenylation sites [p(A)L]. (B) Bar chart of CAT activity per 103 HeLa cells of deletion mutant constructs assayed in the presence of [3H]chloramphenicol, as the means plus standard deviations of duplicate transfections from three separate experiments. (C) Splice site mutant constructs. Line 1, wild-type sequence of the 5′ NRE in the expression plasmid pCAT5′. Boxed sequences, weak consensus 5′ splice sites (1 to 4). The shill marks the position of the exon-intron boundary. Line 2, pCATwt2SD contains a group of mutations (boldface) that convert 5′ splice site 2 to a perfect consensus 5′ splice site sequence. Line 3, pCATmut1-4SD contains inactivating point mutations (boldface) in all four 5′ splice sites; pCAT5′mut1-4SD also lacks the GU-rich 3′ NRE. Line 4, pCAT5′mut1-3SD contains inactivating point mutations in three 5′ splice sites (boldface) but retains wild-type sequence at splice site 4. Lines 5 to 8, pCAT5′mut1SD, pCAT5′mut2SD, pCAT5′mut3SD, and pCAT5′mut4SD contain inactivating point mutations (boldface) in splice sites 1 to 4, respectively. (D) Bar chart of CAT activity per 103 HeLa cells of 5′ splice site mutant constructs assayed in the presence of [3H]chloramphenicol, as the means plus standard deviations of duplicate transfections from three separate experiments.
FIG. 3.
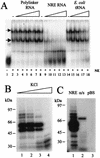
Binding of protein to the NRE is specific and occurs with high binding affinity. (A) Specific competition EMSA. A 32P-labeled NRE probe (1.5 pmol) was incubated with HeLa cell nuclear extracts in the presence of specific or nonspecific competitor RNA. Lane 1, no extract; lane 2, no competitor; lanes 3 to 7, 1- to 16-fold molar excess of nonspecific competitor, i.e., 1.5 to 24 pmol of in vitro-transcribed, unlabeled pBluescript KS(+) polylinker RNA; lane 8, no competitor; lanes 9 to 13, 1- to 16-fold molar excess of specific competitor, i.e., 1.5 to 24 pmol of in vitro-transcribed NRE RNA; lane 14, no competitor RNA; lanes 15 to 18, nonspecific competitor, i.e., 500 ng to 2 μg of E. coli tRNA. Arrows, RNA-protein complexes; NE, HeLa cell nuclear extracts. (B) A 32P-labeled NRE probe was UV cross-linked to HeLa cell nuclear extracts in the presence of various concentrations of KCl (60, 120, 250, and 500 mM [lanes 1 to 4, respectively]). (C) 32P-labeled sense NRE (lane 1), antisense NRE (lane 2), and pBluescript polylinker probes (lane 3) were UV cross-linked to HeLa cell nuclear extracts in 60 mM KCl. In panels B and C cross-linked protein-RNA samples were RNase digested and fractionated on SDS-polyacrylamide gels, dried, and subjected to autoradiography.
FIG.4.
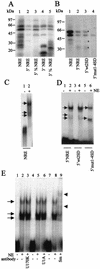
UV cross-linking and EMSA with mutant NRE probes and HeLa cell nuclear extracts. (A) UV cross-linking of 32P-labeled wild-type and deletion mutant NRE probes to HeLa cell nuclear extracts. Lane 1, NRE; lane 2, 5′ NRE; lane 3, 5′[3/4] NRE; lane 4, 3′ NRE; lane 5, 3′[3/4] NRE. (B) UV cross-linking of [32P]rATP-labeled whole-NRE, 5′ NRE, and 5′ splice site mutant probes to HeLa cell nuclear extracts. Lane 1, whole-NRE probe; lane 2, 5′ NRE probe; lane 3, 5′wt2SD probe; lane 4, 5′mut1-4SD probe. Asterisks, proteins binding to 5′wt2SD and 5′ NRE but not to 5′mut1-4SD. (C) EMSA with 32P-labeled whole-NRE probe and HeLa cell nuclear extracts. Lane 1, no nuclear extract; lane 2, with HeLa cell nuclear extract. Arrows, RNA-protein complexes. (D) EMSA with 32P-labeled short (49-nt) 5′ splice site mutant NRE probes and HeLa cell nuclear extracts. Lanes 1 and 2, 5′ NRE probe; lanes 3 and 4, 5′wt2SD probe; lanes 5 and 6, 5′mut1-4SD probe. Arrows, RNA-protein complexes; NE, HeLa cell nuclear extracts. (E) EMSA with 32P-labeled whole NRE probe and HeLa cell nuclear extracts preincubated with various antibodies. Lanes 1, 4, and 7, no nuclear extract and no antibody; lanes 2, 5, and 8, nuclear extract (NE) and no antibody; lane 3, nuclear extract with anti-U1 70K antibody; lane 6, nuclear extract with anti-U1A antibody; lane 9, nuclear extract with anti-Sm antibody. Arrows, RNA-protein complexes. Arrowheads mark the positions of shifted complexes.
FIG. 5.
Affinity purification and identification of proteins and RNA that interact with NRE and splice site mutant RNAs. (A) Western blotting with a polyclonal U1A antiserum. Lanes 1 and 9, NRE-binding proteins isolated from HeLa cell nuclear extracts; lane 2, wt2SD-binding proteins; lane 3, mut1-4SD-binding proteins; lane 4, proteins purified with beads alone; lane 5, 20 μg of HeLa cell nuclear extracts; lane 6, 5′ NRE-binding proteins; lane 7, 5′wt2SD-binding proteins; lane 8, 5′mut1-4SD-binding proteins (the exposure of lanes 6 to 8 is 10 times longer than that of lanes 1 to 5); lane 10, 3′ NRE-binding proteins; lane 11, 2 μg of HeLa nuclear extracts. (B) Western blotting with the Y12 Sm protein antibody. Lane 1, NRE-binding proteins; lane 2, wt2SD-binding proteins; lane 3, mut1-4SD-binding proteins; lane 4, proteins purified with beads alone; lane 5, 20 μg of HeLa nuclear extracts. (C) Western blotting with the U1 70K antibody. Lanes are as in panel B. (D) Western blotting of proteins coimmunoprecipitated with anti-Sm antibody from HeLa cell nuclear extracts. Lane 1, nuclear extract alone; lane 2, nuclear extract incubated with Sm antibody Y12-bound protein A-Sepharose beads; lane 3, nuclear extract incubated with protein A-Sepharose beads without anti-Sm antibody. The top panel was probed with anti-U1 70K antibody, the middle panel was probed with anti-U1A antibody, and the bottom panel was probed with anti-Sm antibody. (E) Ethidium bromide-stained agarose gel of reverse transcription-PCR-amplified U1 snRNA affinity purified on NRE RNA. Lane 1, 1-kb DNA ladder (Invitrogen); lane 2, no cDNA added; lane 3, NRE affinity-purified U1 snRNA cDNA; lane 4, NRE affinity-purified RNA, no reverse transcriptase; lane 5, affinity-purified cDNA from beads alone; lane 6, affinity-purified cDNA from HeLa cell nuclear extract.
Similar articles
- Human papillomavirus type 16 late gene expression is regulated by cellular RNA processing factors in response to epithelial differentiation.
Cumming SA, Cheun-Im T, Milligan SG, Graham SV. Cumming SA, et al. Biochem Soc Trans. 2008 Jun;36(Pt 3):522-4. doi: 10.1042/BST0360522. Biochem Soc Trans. 2008. PMID: 18481996 Free PMC article. Review. - Sequences homologous to 5' splice sites are required for the inhibitory activity of papillomavirus late 3' untranslated regions.
Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC. Furth PA, et al. Mol Cell Biol. 1994 Aug;14(8):5278-89. doi: 10.1128/mcb.14.8.5278-5289.1994. Mol Cell Biol. 1994. PMID: 8035806 Free PMC article. - The regulatory element in the 3'-untranslated region of human papillomavirus 16 inhibits expression by binding CUG-binding protein 1.
Goraczniak R, Gunderson SI. Goraczniak R, et al. J Biol Chem. 2008 Jan 25;283(4):2286-96. doi: 10.1074/jbc.M708789200. Epub 2007 Nov 27. J Biol Chem. 2008. PMID: 18042543 - The human papillomavirus type 16 negative regulatory RNA element interacts with three proteins that act at different posttranscriptional levels.
Koffa MD, Graham SV, Takagaki Y, Manley JL, Clements JB. Koffa MD, et al. Proc Natl Acad Sci U S A. 2000 Apr 25;97(9):4677-82. doi: 10.1073/pnas.070049097. Proc Natl Acad Sci U S A. 2000. PMID: 10781073 Free PMC article. - A novel role of U1 snRNP: Splice site selection from a distance.
Singh RN, Singh NN. Singh RN, et al. Biochim Biophys Acta Gene Regul Mech. 2019 Jun;1862(6):634-642. doi: 10.1016/j.bbagrm.2019.04.004. Epub 2019 Apr 28. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 31042550 Free PMC article. Review.
Cited by
- Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF.
Mole S, Milligan SG, Graham SV. Mole S, et al. J Virol. 2009 Jan;83(1):357-67. doi: 10.1128/JVI.01414-08. Epub 2008 Oct 22. J Virol. 2009. PMID: 18945764 Free PMC article. - The RNA stability regulator HuR regulates L1 protein expression in vivo in differentiating cervical epithelial cells.
Cumming SA, Chuen-Im T, Zhang J, Graham SV. Cumming SA, et al. Virology. 2009 Jan 5;383(1):142-9. doi: 10.1016/j.virol.2008.10.003. Epub 2008 Nov 4. Virology. 2009. PMID: 18986664 Free PMC article. - Identifying regulatory elements and their RNA-binding proteins in the 3' untranslated regions of papillomavirus late mRNAs.
Iamborwornkun N, Kitkumthorn N, Stevenson A, Kirk A, Graham SV, Chuen-Im T. Iamborwornkun N, et al. Biomed Rep. 2024 Jul 1;21(2):125. doi: 10.3892/br.2024.1813. eCollection 2024 Aug. Biomed Rep. 2024. PMID: 39006509 Free PMC article. - A 57-nucleotide upstream early polyadenylation element in human papillomavirus type 16 interacts with hFip1, CstF-64, hnRNP C1/C2, and polypyrimidine tract binding protein.
Zhao X, Oberg D, Rush M, Fay J, Lambkin H, Schwartz S. Zhao X, et al. J Virol. 2005 Apr;79(7):4270-88. doi: 10.1128/JVI.79.7.4270-4288.2005. J Virol. 2005. PMID: 15767428 Free PMC article. - Human papillomavirus type 16 late gene expression is regulated by cellular RNA processing factors in response to epithelial differentiation.
Cumming SA, Cheun-Im T, Milligan SG, Graham SV. Cumming SA, et al. Biochem Soc Trans. 2008 Jun;36(Pt 3):522-4. doi: 10.1042/BST0360522. Biochem Soc Trans. 2008. PMID: 18481996 Free PMC article. Review.
References
- Black, D. J., R. Chan, H. Min, J. Wang, and L. Bell. 1998. The electrophoretic mobility shift assay for RNA binding proteins, p. 109-136. In C. W. J. Smith (ed.), RNA-protein interactions: a practical approach. Oxford University Press, Oxford, United Kingdom.
- Collier, B., L. Goobar-Larsson, M. Sokolowski, and S. Schwartz. 1998. Translational inhibition of human papillomavirus type 16 L2 mRNA mediated through interaction with heterogeneous ribonucleoprotein K and poly(rC) binding proteins 1 and 2. J. Biol. Chem. 273:22648-22656. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials