TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells - PubMed (original) (raw)
TACE cleavage of proamphiregulin regulates GPCR-induced proliferation and motility of cancer cells
Andreas Gschwind et al. EMBO J. 2003.
Abstract
Communication between G protein-coupled receptor (GPCR) and epidermal growth factor receptor (EGFR) signalling systems involves cell surface proteolysis of EGF-like precursors. The underlying mechanisms of EGFR signal transactivation pathways, however, are largely unknown. We demonstrate that in squamous cell carcinoma cells, stimulation with the GPCR agonists LPA or carbachol specifically results in metalloprotease cleavage and release of amphiregulin (AR). Moreover, AR gene silencing by siRNA or inhibition of AR biological activity by neutralizing antibodies and heparin prevents GPCR-induced EGFR tyrosine phosphorylation, downstream mitogenic signalling events, cell proliferation, migration and activation of the survival mediator Akt/PKB. Therefore, despite some functional redundancy among EGF family ligands, the present study reveals a distinct and essential role for AR in GPCR-triggered cellular responses. Furthermore, we present evidence that blockade of the metalloprotease-disintegrin tumour necrosis factor-alpha-converting enzyme (TACE) by the tissue inhibitor of metalloprotease-3, a dominant-negative TACE mutant or RNA interference suppresses GPCR-stimulated AR release, EGFR activation and downstream events. Thus, TACE can function as an effector of GPCR-mediated signalling and represents a key element of the cellular receptor cross-talk network.
Figures
Fig. 1. EGFR signal transactivation requires metalloprotease activity and the extracellular domain of the EGFR. SCC-9 cells were pre-incubated with marimastat (BB2516, 10 µM; 20 min), anti-EGFR antibody ICR-3R (20 µg/ml; 60 min) or PTX (100 ng/ml; 18 h) and treated with LPA (10 µM), carbachol (Car, 1 mM), EGF (7.5 ng/ml) or pervanadate (PV, 1 mM) for 3 min. Following immunoprecipitation (IP) of cell extracts with anti-EGFR antibody, proteins were immunoblotted (IB) with anti-phosphotyrosine antibody and re-probed with anti-EGFR antibody.
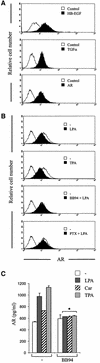
Fig. 2. GPCR stimulation results in metalloprotease-dependent cleavage and release of AR at the cell surface. (A) Flow cytometric analysis of EGF-like precursor expression. SCC-9 cells were collected and stained for surface HB-EGF, TGF-α or AR and analysed by flow cytometry. Control cells were labelled with FITC-conjugated secondary antibody alone. (B) LPA-induced proteolytic processing of proAR. SCC-9 cells were pre-incubated with batimastat (BB94; 10 µM) or PTX and stimulated with LPA or TPA (1 µM) for 5 min. Cells were collected and analysed for cell surface AR density by flow cytometry. (C) GPCR-induced proteolytic release of AR. SCC-9 cells were pre-incubated with batimastat or vehicle followed by stimulation with agonists as indicated for 120 min. Conditioned medium was collected and analysed for total AR amount by ELISA. Each bar is the average of triplicate values (mean ± SD). *P < 0.03 for the difference between agonists versus BB94 + agonists.
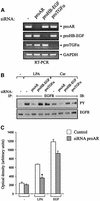
Fig. 3. Effect of proAR siRNA on GPCR-induced EGFR activation and cell migration. (A) Blockade of EGF-like growth factor precursor expression by RNA interference (RNAi). SCC-9 cells were transfected with siRNA for proAR, proHB-EGF or proTGF-α, cultured for 2 days and analysed for gene expression by RT–PCR as indicated or (B) stimulated with LPA or carbachol and assayed for EGFR tyrosine phosphorylation content. (C) Requirement of AR for LPA-induced cell migration. SiRNA-transfected SCC-9 cells were analysed for transwell migration toward fibronectin as chemoattractant. Each bar is the average of quadruplicate values (mean ± SD). *P < 0.001 for control siRNA + LPA versus proAR siRNA + LPA.
Fig. 4. Inhibition of AR bioactivity by anti-AR neutralizing antibodies and heparin abrogates EGFR tyrosine phorphorylation, mitogenic signalling events, activation of Akt/PKB and cell proliferation by GPCR ligands. (A) SCC-9 cells were pre-treated with anti-AR antibody (αAR Ab; 50 µg/ml, 60 min) or heparin (100 ng/ml, 15 min), and stimulated for 3 min (EGFR, upper panel) or 5 min (SHC, lower panel) as indicated. Precipitated EGFR and SHC were immunoblotted with anti-phosphotyrosine antibody followed by reprobing of the same filters with anti-EGFR and anti-SHC antibody, respectively. (B) Association of Grb2 with SHC in vitro. SCC-9 cells were pre-incubated with inhibitors and stimulated for 5 min as indicated. Lysates were incubated with GST–Grb-2 fusion protein or GST alone. Proteins were immunoblotted with monoclonal anti-SHC antibody. (C) AR is required for GPCR-induced ERK/MAPK activation and Akt/PKB phosphorylation. SCC-9 or SCC-15 cells were pre-incubated with inhibitors and stimulated for 7 min. Phosphorylated ERK1/2 was detected by immunoblotting total lysates with anti-phospho-ERK antibody. The same filters were re-probed with anti-ERK antibody. Quantitative analysis of ERK phosphorylation from three independent experiments (mean ± SD). *P < 0.05 for the difference between LPA versus inhibitors + LPA. Stimulation of Akt/PKB. Cell lysates were immunoblotted with anti-phospho-Akt/PKB antibody followed by reprobing of the same filters with anti-Akt/PKB antibody. (D) Effect of PI3K and EGFR inhibition on GPCR-induced Akt/PKB phosphorylation. Quiescent SCC-9 cells were pre-treated with wortmannin (WM, 100 nM), LY294002 (100 µM), AG1478 (250 nM) or vehicle for 30 min and stimulated with LPA or carbachol for 15 min. After lysis, activated Akt/PKB was detected by immunoblotting of total lysates with polyclonal anti-phospho-Akt/PKB (P-Akt) antibody, followed by reprobing of the same filter with polyclonal anti-Akt/PKB (Akt) antibody. (E) Effect of AR inhibition on LPA-induced DNA synthesis. SCC-15 cells were treated with inhibitors as indicated and incubated in the presence or absence of ligands (LPA; AR, 10 ng/ml) for 18 h. Cells were then pulse labelled with [3H]thymidine, and thymidine incorporation was measured by liquid scintillation counting. Quantitative analysis from three independent experiments (mean ± SD). *P < 0.001 for LPA versus inhibitors + LPA.
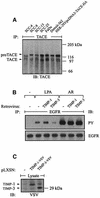
Fig. 5. Expression of TACE in HNSCC cell lines and effect of Timp-1 and Timp-3 expression on the EGFR transactivation signal. (A) TACE was immunoprecipitated from lysates of HNSCC cells with monoclonal TACE antibody. HEK-293 cells transfected with human TACE cDNA served as positive control. (B) Timp-3, but not Timp-1, inhibits EGFR signal transactivation. SCC-9 cells were infected with retrovirus encoding human Timp-1 or Timp-3. EGFR activation was determined by immunoblot after stimulation with agonists as indicated. (C) Expression of Timp-1 and TIMP-3 carrying a C-terminal VSV tag was confirmed by immunoblotting total cell lysates with anti-VSV antibody.
Fig. 6. Dominant-negative TACE suppresses GPCR-induced AR release and EGFR signal transactivation. (A) Expression of wild-type (Wt 17) and dominant-negative TACE (ΔMP 17) in SCC-9 cells after retroviral gene transfer. Total lysates were immunoblotted with polyclonal anti-TACE antibody. (B) Dominant-negative TACE (ΔMP 17) abrogates LPA-induced proAR cleavage and (C) AR release into cell culture medium as determined by flow cytometric analysis and AR ELISA, respectively. (D) Effect of wild-type (Wt 17), dominant-negative TACE (ΔMP 17) and dominant-negative ADAM12 (ΔMP 12) on GPCR-stimulated EGFR signal transactivation. (E) Effect of dominant-negative ADAM10 (ΔMP 10), ADAM12 (ΔMP 12), ADAM15 (ΔMP 15) and TACE (ΔMP 17) on GPCR-stimulated EGFR signal transactivation. (F) Expression of dominant-negative ADAM mutants carrying a C-terminal HA tag in SCC-9 cells after retroviral gene transfer. Total lysates were immunoblotted with anti-HA antibody.
Fig. 7. TACE siRNA inhibits EGFR signal transmission and cell migration by GPCR agonists. (A and B) TACE siRNA blocks endogenous TACE expression. SCC-9 cells were transfected with TACE or ADAM12 siRNA. Gene expression was analysed by (A) RT–PCR or (B) immunoblot with polyclonal anti-TACE antibody. (C) Knockdown of TACE results in accumulation of proAR at the cell surface. siRNA-transfected SCC-9 cells were analysed for AR cell surface content by FACS. (D) EGFR signal transmission upon GPCR activation requires TACE. SCC-9 cells were transfected with siRNA and stimulated with agonists as indicated. Activation of EGFR, SHC, ERK and Akt was determined as described above. (E) Squamous cancer cell motility in response to LPA depends on TACE. siRNA-transfected SCC-9 cells were treated with LPA or AR and analysed in a transwell migration assay.
Similar articles
- GPCR-induced migration of breast carcinoma cells depends on both EGFR signal transactivation and EGFR-independent pathways.
Hart S, Fischer OM, Prenzel N, Zwick-Wallasch E, Schneider M, Hennighausen L, Ullrich A. Hart S, et al. Biol Chem. 2005 Sep;386(9):845-55. doi: 10.1515/BC.2005.099. Biol Chem. 2005. PMID: 16164409 - TACE/ADAM17 processing of EGFR ligands indicates a role as a physiological convertase.
Lee DC, Sunnarborg SW, Hinkle CL, Myers TJ, Stevenson MY, Russell WE, Castner BJ, Gerhart MJ, Paxton RJ, Black RA, Chang A, Jackson LF. Lee DC, et al. Ann N Y Acad Sci. 2003 May;995:22-38. doi: 10.1111/j.1749-6632.2003.tb03207.x. Ann N Y Acad Sci. 2003. PMID: 12814936 Review. - ADAM-mediated ectodomain shedding of HB-EGF in receptor cross-talk.
Higashiyama S, Nanba D. Higashiyama S, et al. Biochim Biophys Acta. 2005 Aug 1;1751(1):110-7. doi: 10.1016/j.bbapap.2004.11.009. Epub 2004 Dec 8. Biochim Biophys Acta. 2005. PMID: 16054021 Review.
Cited by
- ADAM-Mediated Signalling Pathways in Gastrointestinal Cancer Formation.
Schumacher N, Rose-John S, Schmidt-Arras D. Schumacher N, et al. Int J Mol Sci. 2020 Jul 20;21(14):5133. doi: 10.3390/ijms21145133. Int J Mol Sci. 2020. PMID: 32698506 Free PMC article. Review. - Lysophosphatidic acid induces tumor necrosis factor-alpha to regulate a pro-inflammatory cytokine network in ovarian cancer.
Wang W, Wu J, Mukherjee A, He T, Wang XY, Ma Y, Fang X. Wang W, et al. FASEB J. 2020 Oct;34(10):13935-13948. doi: 10.1096/fj.202001136R. Epub 2020 Aug 26. FASEB J. 2020. PMID: 32851734 Free PMC article. - ADAM10 mediates ectopic proximal tubule development and renal fibrosis through Notch signalling.
Li B, Zhu C, Dong L, Qin J, Xiang W, Davidson AJ, Feng S, Wang Y, Shen X, Weng C, Wang C, Zhu T, Teng L, Wang J, Englert C, Chen J, Jiang H. Li B, et al. J Pathol. 2020 Nov;252(3):274-289. doi: 10.1002/path.5517. Epub 2020 Sep 24. J Pathol. 2020. PMID: 32715474 Free PMC article. - Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands.
Sahin U, Weskamp G, Kelly K, Zhou HM, Higashiyama S, Peschon J, Hartmann D, Saftig P, Blobel CP. Sahin U, et al. J Cell Biol. 2004 Mar 1;164(5):769-79. doi: 10.1083/jcb.200307137. J Cell Biol. 2004. PMID: 14993236 Free PMC article. - Adhesion-mediated squamous cell carcinoma survival through ligand-independent activation of epidermal growth factor receptor.
Shen X, Kramer RH. Shen X, et al. Am J Pathol. 2004 Oct;165(4):1315-29. doi: 10.1016/S0002-9440(10)63390-1. Am J Pathol. 2004. PMID: 15466396 Free PMC article.
References
- Amour A. et al. (1998) TNF-α converting enzyme (TACE) is inhibited by TIMP-3. FEBS Lett., 435, 39–44. - PubMed
- Amour A., Knight,C.G., Webster,A., Slocombe,P.M., Stephens,P.E., Knauper,V., Docherty,A.J. and Murphy,G. (2000) The in vitro activity of ADAM-10 is inhibited by TIMP-1 and TIMP-3. FEBS Lett., 473, 275–279. - PubMed
- Asakura M. et al. (2002) Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat. Med., 8, 35–40. - PubMed
- Bar-Sagi D. and Hall,A. (2000) Ras and Rho GTPases: a family reunion. Cell, 103, 227–238. - PubMed
- Brown C.L., Coffey,R.J. and Dempsey,P.J. (2001) The proamphiregulin cytoplasmic domain is required for basolateral sorting, but is not essential for constitutive or stimulus-induced processing in polarized Madin–Darby canine kidney cells. J. Biol. Chem., 276, 29538–29549. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous