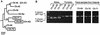Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii) - PubMed (original) (raw)
. 2003 Jul;77(13):7545-62.
doi: 10.1128/jvi.77.13.7545-7562.2003.
Magdalena Lukasik, Shadrack Kamenya, Yingying Li, Frederic Bibollet-Ruche, Elizabeth Bailes, Martin N Muller, Melissa Emery, David A Goldenberg, Jeremiah S Lwanga, Ahidjo Ayouba, Eric Nerrienet, Harold M McClure, Jonathan L Heeney, David P Watts, Anne E Pusey, D Anthony Collins, Richard W Wrangham, Jane Goodall, John F Y Brookfield, Paul M Sharp, George M Shaw, Beatrice H Hahn
Affiliations
- PMID: 12805455
- PMCID: PMC164799
- DOI: 10.1128/jvi.77.13.7545-7562.2003
Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii)
Mario L Santiago et al. J Virol. 2003 Jul.
Abstract
Simian immunodeficiency virus of chimpanzees (SIVcpz) is the immediate precursor to human immunodeficiency virus type 1 (HIV-1), yet remarkably, the distribution and prevalence of SIVcpz in wild ape populations are unknown. Studies of SIVcpz infection rates in wild chimpanzees are complicated by the species' endangered status and by its geographic location in remote areas of sub-Saharan Africa. We have developed sensitive and specific urine and fecal tests for SIVcpz antibody and virion RNA (vRNA) detection and describe herein the first comprehensive prevalence study of SIVcpz infection in five wild Pan troglodytes schweinfurthii communities in east Africa. In Kibale National Park in Uganda, 31 (of 52) members of the Kanyawara community and 39 (of approximately 145) members of the Ngogo community were studied; none were found to be positive for SIVcpz infection. In Gombe National Park in Tanzania, 15 (of 20) members of the Mitumba community, 51 (of 55) members of the Kasekela community, and at least 10 (of approximately 20) members of the Kalande community were studied. Seven individuals were SIVcpz antibody and/or vRNA positive, and two others had indeterminate antibody results. Based on assay sensitivities and the numbers and types of specimens analyzed, we estimated the prevalence of SIVcpz infection to be 17% in Mitumba (95% confidence interval, 10 to 40%), 5% in Kasekela (95% confidence interval, 4 to 7%), and 30% in Kalande (95% confidence interval, 15 to 60%). For Gombe as a whole, the SIVcpz prevalence was estimated to be 13% (95% confidence interval, 7 to 25%). SIVcpz infection was confirmed in five chimpanzees by PCR amplification of partial pol and gp41/nef sequences which revealed a diverse group of viruses that formed a monophyletic lineage within the SIVcpzPts radiation. Although none of the 70 Kibale chimpanzees tested SIVcpz positive, we estimated the likelihood that a 10% or higher prevalence existed but went undetected because of sampling and assay limitations; this possibility was ruled out with 95% certainty. These results indicate that SIVcpz is unevenly distributed among P. t. schweinfurthii in east Africa, with foci or "hot spots" of SIVcpz endemicity in some communities and rare or absent infection in others. This situation contrasts with that for smaller monkey species, in which infection rates by related SIVs are generally much higher and more uniform among different groups and populations. The basis for the wide variability in SIVcpz infection rates in east African apes and the important question of SIVcpz prevalence in west central African chimpanzees (Pan troglodytes troglodytes) remain to be elucidated.
Figures
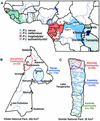
FIG.1.
Location of wild chimpanzee study sites. (A) The locations of Kibale (Uganda) and Gombe (Tanzania) National Parks (indicated by asterisks) are shown in relation to the range of the common chimpanzees in equatorial Africa (Copyright © 2001, Smithsonian Institution. Adapted from reference with permission). The four recognized chimpanzee subspecies are color coded. International borders (black lines) and major rivers (blue lines) are shown. Uganda and Tanzania are highlighted. (B) Map of Kibale National Park, indicating the approximate ranges of the Kanyawara (red) and Ngogo (blue) communities. The two communities are not adjacent, thus limiting direct contact between members. Black dots denote ranger outposts. (C) Map of Gombe National Park, indicating the approximate ranges of the northern Mitumba (red), the main Kasekela (blue), and the nonhabituated southern Kalande community (green). Gombe National Park is bordered by Lake Tanganyika to the east, and the rift escarpment (>1,500 m) to the west. Interactions between members of the different communities have been observed in regions of overlap.
FIG. 2.
Detection of HIV-1- and SIVcpz-specific antibodies in chimpanzee fecal and urine samples. Fecal (A) and urine (B) specimens from captive chimpanzees chronically infected with either HIV-1 or SIVcpz, and specimens from uninfected controls, were tested by ECL Western immunoblot analysis for the presence of virus-specific antibodies. Chimpanzees are identified by code number (additional information for each individual is provided in Table 1); Ch-No and Ch-Ni are infected with the highly divergent SIVcpzANT strain (28, 55). Serial samples from the same individual are identified in parentheses. Molecular weights of HIV-1-specific proteins are indicated. The banding pattern of plasma from an HIV-1-infected human (analyzed at dilutions of 1:10,000 and 1:100,000) is shown for positive control.
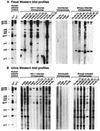
FIG. 3.
Western blot profiles of urine samples from wild-living chimpanzees. Representative Western immunoblots of urine samples collected in Kanyawara, Kasekela, and Mitumba are shown; collection dates are indicated (Tables 3 to 5 show details on the particular chimpanzees analyzed). Molecular weights of HIV-1-specific proteins are indicated. The banding pattern of plasma from an HIV-1-infected human (analyzed at dilutions of 1:100,000) is shown for positive control.
FIG. 4.
Subspecies and sex determination of three SIVcpz-infected, nonhabituated chimpanzees from Kalande. (A) Phylogenetic tree of mitochondrial DNA sequences. D-loop sequences (498 bp) amplified from three SIVcpz vRNA-positive fecal specimens (boxed) from Kalande were compared with mtDNA sequences from other eastern chimpanzees (P. t. schweinfurthii). The mtDNA sequence for Ch-No has been reported previously (18). Sequences for Ch-06, Ch-22, Ch-45, Ch-37, Ch-30, and Ch-Ni have been determined in this study. The tree was obtained by maximum likelihood analysis of 377 gap-stripped sites by using the REV + Gamma substitution model implemented in BASEML from the PAML package (PAML, software version 3.0b; University College London, London, United Kingdom) testing 48 tree topologies obtained from NUCML from the MOLPHY package (program package for MOLecular PHYlogenetics, software version 2.2; Institute of Statistical Mathematics, 4-6-7 Minami-Azabu, Minato-ku, Tokyo 106, Japan). Asterisks denote clades to the right found in at least 70% of bootstrapped replicates analyzed by the neighbor-joining method implemented in CLUSTAL W (53) using Kimura's two-parameter correction (27). The three Kalande sequences (boxed) represent different haplotypes within the P. t. schweinfurthii radiation, indicating that the samples came from three different individuals. Ch-30 and Ch-37 (in parentheses) have haplotypes identical to those of Ch-64 and Ch-45, respectively. The tree is rooted on sequences from the subspecies P. t. verus and P. t. vellerosus. (B) Sex determination of Ch-64, Ch-70, and Ch-71 from Kalande. A region of the amelogenin gene, known to contain a 189-bp deletion near the centromere of the Y, but not the X, chromosome, was amplified (16). Fecal DNA from known male (Ch-06, Ch-05, and Ch-16) and female (Ch-09, Ch-25, and Ch-01) chimpanzees from Kasekela was analyzed for control. Sex determination for Ch-64, Ch-70, and Ch-71 was performed in duplicate.
FIG. 5.
Phylogenetic analyses of SIVcpz in Gombe. Five different SIVcpz strains identified in Mitumba, Kasekela, and Kalande were analyzed in two different genomic regions, a 286-bp pol and 564-bp gp41/nef fragment (after gap stripping). The newly derived sequences TAN2, TAN3, TAN4, and TAN5 were compared with TAN1 sequences from different time points (Table 5) as well as reference sequences for HIV-1 M subtype A; U455 (GenBank accession number M62320), subtype B; LAI (K02013), subtype D; ELI (K03454), HIV-1 N; YBF30 (AJ006022), HIV-1 O; MVP5180 (L20771), and ANT70C (L20587), SIVcpz; CAM3 (AF115393), CAM5 (AJ271369), US (AF103818), GAB1 (X52154), and GAB2 (F. Bibollet-Ruche and B. H. Hahn, unpublished data), and ANT (U42720). The trees were obtained by the neighbor-joining method (47) implemented in CLUSTAL W (53) by using Kimura's two-parameter correction (27) and 1,000 bootstrapped replicates. Asterisks indicate that the clades to the right were found in at least 80% of bootstrapped replicates.
Similar articles
- Origins of HIV and the AIDS pandemic.
Sharp PM, Hahn BH. Sharp PM, et al. Cold Spring Harb Perspect Med. 2011 Sep;1(1):a006841. doi: 10.1101/cshperspect.a006841. Cold Spring Harb Perspect Med. 2011. PMID: 22229120 Free PMC article. Review. - Impact of simian immunodeficiency virus infection on chimpanzee population dynamics.
Rudicell RS, Holland Jones J, Wroblewski EE, Learn GH, Li Y, Robertson JD, Greengrass E, Grossmann F, Kamenya S, Pintea L, Mjungu DC, Lonsdorf EV, Mosser A, Lehman C, Collins DA, Keele BF, Goodall J, Hahn BH, Pusey AE, Wilson ML. Rudicell RS, et al. PLoS Pathog. 2010 Sep 23;6(9):e1001116. doi: 10.1371/journal.ppat.1001116. PLoS Pathog. 2010. PMID: 20886099 Free PMC article. - High prevalence of simian immunodeficiency virus infection in a community of savanna chimpanzees.
Rudicell RS, Piel AK, Stewart F, Moore DL, Learn GH, Li Y, Takehisa J, Pintea L, Shaw GM, Moore J, Sharp PM, Hahn BH. Rudicell RS, et al. J Virol. 2011 Oct;85(19):9918-28. doi: 10.1128/JVI.05475-11. Epub 2011 Jul 20. J Virol. 2011. PMID: 21775446 Free PMC article. - Amplification of a complete simian immunodeficiency virus genome from fecal RNA of a wild chimpanzee.
Santiago ML, Bibollet-Ruche F, Bailes E, Kamenya S, Muller MN, Lukasik M, Pusey AE, Collins DA, Wrangham RW, Goodall J, Shaw GM, Sharp PM, Hahn BH. Santiago ML, et al. J Virol. 2003 Feb;77(3):2233-42. doi: 10.1128/jvi.77.3.2233-2242.2003. J Virol. 2003. PMID: 12525658 Free PMC article. - Simian retroviruses in African apes.
Peeters M, Delaporte E. Peeters M, et al. Clin Microbiol Infect. 2012 Jun;18(6):514-20. doi: 10.1111/j.1469-0691.2012.03843.x. Epub 2012 Apr 20. Clin Microbiol Infect. 2012. PMID: 22515409 Review.
Cited by
- Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir.
Li Y, Ndjango JB, Learn GH, Ramirez MA, Keele BF, Bibollet-Ruche F, Liu W, Easlick JL, Decker JM, Rudicell RS, Inogwabini BI, Ahuka-Mundeke S, Leendertz FH, Reynolds V, Muller MN, Chancellor RL, Rundus AS, Simmons N, Worobey M, Shaw GM, Peeters M, Sharp PM, Hahn BH. Li Y, et al. J Virol. 2012 Oct;86(19):10776-91. doi: 10.1128/JVI.01498-12. Epub 2012 Jul 25. J Virol. 2012. PMID: 22837215 Free PMC article. - The impact of genetic adaptation on chimpanzee subspecies differentiation.
Schmidt JM, de Manuel M, Marques-Bonet T, Castellano S, Andrés AM. Schmidt JM, et al. PLoS Genet. 2019 Nov 25;15(11):e1008485. doi: 10.1371/journal.pgen.1008485. eCollection 2019 Nov. PLoS Genet. 2019. PMID: 31765391 Free PMC article. - Origins of HIV and the AIDS pandemic.
Sharp PM, Hahn BH. Sharp PM, et al. Cold Spring Harb Perspect Med. 2011 Sep;1(1):a006841. doi: 10.1101/cshperspect.a006841. Cold Spring Harb Perspect Med. 2011. PMID: 22229120 Free PMC article. Review. - Field immobilization for treatment of an unknown illness in a wild chimpanzee (Pan troglodytes schweinfurthii) at Gombe National Park, Tanzania: findings, challenges, and lessons learned.
Lonsdorf E, Travis D, Ssuna R, Lantz E, Wilson M, Gamble K, Terio K, Leendertz F, Ehlers B, Keele B, Hahn B, Gillespie T, Pond J, Raphael J, Collins A. Lonsdorf E, et al. Primates. 2014 Jan;55(1):89-99. doi: 10.1007/s10329-013-0372-4. Epub 2013 Jul 20. Primates. 2014. PMID: 23872909 Free PMC article. - Transmission of simian immunodeficiency virus SIVcpz and the evolution of infection in the presence and absence of concurrent human immunodeficiency virus type 1 infection in chimpanzees.
Heeney JL, Rutjens E, Verschoor EJ, Niphuis H, ten Haaft P, Rouse S, McClure H, Balla-Jhagjhoorsingh S, Bogers W, Salas M, Cobb K, Kestens L, Davis D, van der Groen G, Courgnaud V, Peeters M, Murthy KK. Heeney JL, et al. J Virol. 2006 Jul;80(14):7208-18. doi: 10.1128/JVI.00382-06. J Virol. 2006. PMID: 16809326 Free PMC article.
References
- Bailes, E., R. R. Chaudhuri, M. L. Santiago, F. Bibollet-Ruche, B. H. Hahn, and P. M. Sharp. 2002. The evolution of primate lentiviruses and the origins of AIDS, p. 65-96. In T. Leitner (ed.), The molecular epidemiology of human viruses. Kluwer Academic Publishers, Norwell, Mass.
- Bailes, E., F. Gao, F. Bibollet-Ruche, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science, in press. - PubMed
- Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsch. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. - PMC - PubMed
- Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77:773-781. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous