Detection of reliable and unexpected protein fold predictions using 3D-Jury - PubMed (original) (raw)
Detection of reliable and unexpected protein fold predictions using 3D-Jury
Krzysztof Ginalski et al. Nucleic Acids Res. 2003.
Abstract
3D-Jury is a fully automated protein structure meta prediction system accessible via the Meta Server interface (http://BioInfo.PL/Meta). This is one of the meta predictors, which have made a dramatic, unprecedented impact on the last CASP-5 experiment. The 3D-Jury is comparable with other meta servers but it has the highest combined specificity and sensitivity. The presented method is also very simple and versatile and can be used to create meta predictions even from sets of models produced by humans. An additional and very important and novel feature of the system is the high correlation between the reported confidence score and the accuracy of the model. The number of correctly predicted residues can be estimated directly from the prediction score. The high reliability of the method enables any biologist to submit a target of interest to the Meta Server and screen with relatively high confidence, whether the target can be predicted by fold recognition methods while being unpredictable using standard approaches like PSI-Blast. This can point to interesting relationships which could have been missed in annotations of proteins or genomes and provide very valuable information for novel scientific discoveries.
Figures
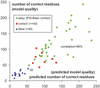
Figure 1
Correlation between predicted and observed quality of models obtained with 3D-Jury. The _x_-axis shows the confidence scores reported by 3D-Jury with default settings for all 55 CASP-5 targets with currently known structure. The servers used by the system for consensus building included: ORFeus (11), SamT02 (12), FFAS03 (13), mGenTHREADER (14), INBGU (15), RAPTOR (16), FUGUE-2 (17) and 3D-PSSM (18). The confidence score equals the average number of corresponding residues (residues that can be superimposed on the model) within a selected set of original models provided by other servers. Surprisingly, this number correlates very well with the accuracy of the 3D-Jury model (the number of residues of the model that can be correctly superimposed on the native structure). The _y_-axis shows the number of C-alpha positions of the model that can be superimposed on the correct structure of the target within 3 Å deviation (correct residues). The correlation between the two values is 86%. Usually, models with 40 or more correct residues are regarded as correct [prediction templates have 90% chance to belong to the same SCOP (19) fold as the targets]. Based on this definition, the points on the plot are divided into 14 false predictions (triangles), eight correct predictions (squares) and 33 correct but easy predictions (rhombs, where PSI-Blast has also generated correct models). 3D-Jury has generated ∼25% more correct predictions than PSI-Blast (PDB-Blast). Only one false prediction (39 correct residues, which is just below the limit for correct predictions) out of 42 has been generated with the default confidence threshold of 50.
Similar articles
- Evaluation of 3D-Jury on CASP7 models.
Kaján L, Rychlewski L. Kaján L, et al. BMC Bioinformatics. 2007 Aug 21;8:304. doi: 10.1186/1471-2105-8-304. BMC Bioinformatics. 2007. PMID: 17711571 Free PMC article. - 3D-Jury: a simple approach to improve protein structure predictions.
Ginalski K, Elofsson A, Fischer D, Rychlewski L. Ginalski K, et al. Bioinformatics. 2003 May 22;19(8):1015-8. doi: 10.1093/bioinformatics/btg124. Bioinformatics. 2003. PMID: 12761065 - Application of 3D-Jury, GRDB, and Verify3D in fold recognition.
von Grotthuss M, Pas J, Wyrwicz L, Ginalski K, Rychlewski L. von Grotthuss M, et al. Proteins. 2003;53 Suppl 6:418-23. doi: 10.1002/prot.10547. Proteins. 2003. PMID: 14579330 - GeneSilico protein structure prediction meta-server.
Kurowski MA, Bujnicki JM. Kurowski MA, et al. Nucleic Acids Res. 2003 Jul 1;31(13):3305-7. doi: 10.1093/nar/gkg557. Nucleic Acids Res. 2003. PMID: 12824313 Free PMC article. - Servers for protein structure prediction.
Fischer D. Fischer D. Curr Opin Struct Biol. 2006 Apr;16(2):178-82. doi: 10.1016/j.sbi.2006.03.004. Epub 2006 Mar 20. Curr Opin Struct Biol. 2006. PMID: 16546376 Review.
Cited by
- Protein structure prediction and analysis using the Robetta server.
Kim DE, Chivian D, Baker D. Kim DE, et al. Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W526-31. doi: 10.1093/nar/gkh468. Nucleic Acids Res. 2004. PMID: 15215442 Free PMC article. - Realm of PD-(D/E)XK nuclease superfamily revisited: detection of novel families with modified transitive meta profile searches.
Knizewski L, Kinch LN, Grishin NV, Rychlewski L, Ginalski K. Knizewski L, et al. BMC Struct Biol. 2007 Jun 20;7:40. doi: 10.1186/1472-6807-7-40. BMC Struct Biol. 2007. PMID: 17584917 Free PMC article. - 24-Epibrassinolide Promotes Fatty Acid Accumulation and the Expression of Related Genes in Styrax tonkinensis Seeds.
Chen C, Chen H, Han C, Liu Z, Yu F, Wu Q. Chen C, et al. Int J Mol Sci. 2022 Aug 10;23(16):8897. doi: 10.3390/ijms23168897. Int J Mol Sci. 2022. PMID: 36012162 Free PMC article. - Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome.
Wlodarski T, Kutner J, Towpik J, Knizewski L, Rychlewski L, Kudlicki A, Rowicka M, Dziembowski A, Ginalski K. Wlodarski T, et al. PLoS One. 2011;6(8):e23168. doi: 10.1371/journal.pone.0023168. Epub 2011 Aug 9. PLoS One. 2011. PMID: 21858014 Free PMC article. - Longin-like folds identified in CHiPS and DUF254 proteins: vesicle trafficking complexes conserved in eukaryotic evolution.
Kinch LN, Grishin NV. Kinch LN, et al. Protein Sci. 2006 Nov;15(11):2669-74. doi: 10.1110/ps.062419006. Protein Sci. 2006. PMID: 17075139 Free PMC article.
References
- Moult J., Fidelis,K., Zemla,A. and Hubbard,T. (2001) Critical assessment of methods of protein structure prediction (CASP): round IV. Proteins (Suppl. 5), 2–7. - PubMed
- Fischer D., Elofsson,A., Rychlewski,L., Pazos,F., Valencia,A., Rost,B., Ortiz,A.R. and Dunbrack,R.L.,Jr (2001) CAFASP2: the second critical assessment of fully automated structure prediction methods. Proteins (Suppl. 5), 171–183. - PubMed
- Ginalski K., Elofsson,A., Fischer,D. and Rychlewski,L. (2003) 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics, in press. - PubMed
- Fischer D. (2003) 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins, in press. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials