Targeted deletion reveals an essential function for the telomere length regulator Trf1 - PubMed (original) (raw)
Targeted deletion reveals an essential function for the telomere length regulator Trf1
Jan Karlseder et al. Mol Cell Biol. 2003 Sep.
Abstract
The human telomeric DNA binding factor TRF1 (hTRF1) and its interacting proteins TIN2, tankyrase 1 and 2, and PINX1 have been implicated in the regulation of telomerase-dependent telomere length maintenance. Here we show that targeted deletion of exon 1 of the mouse gene encoding Trf1 causes early (day 5 to 6 postcoitus) embryonic lethality. The absence of telomerase did not alter the Terf1(ex1Delta/ex1Delta) lethality, indicating that the phenotype was not due to inappropriate telomere elongation by telomerase. Terf1(ex1Delta/ex1Delta) blastocysts had a severe growth defect of the inner cell mass that was accompanied by apoptosis. However, no evidence was found for telomere uncapping causing this cell death; chromosome spreads of Terf1(ex1Delta/ex1Delta) blastocysts did not reveal chromosome end-to-end fusions, and p53 deficiency only briefly delayed Terf1(ex1Delta/ex1Delta) lethality. These data suggest that murine Trf1 has an essential function that is independent of telomere length regulation.
Figures
FIG. 1.
Targeted deletion of exon 1 of the mouse Terf1 locus. (A) Schematics of the targeting construct (top line), wild-type locus (second line), and targeted locus (third line) are shown. The bottom line shows the _Xho_I/_Hin_dIII and _Xho_I fragments used for genotyping. X, _Xho_I; E, _Eco_RI; H, _Hin_dIII. Positions of PCR primers used for genotyping are indicated by half arrows. Primers are as follows: C, shared by the wild-type and the targeted allele; wt, wild-type allele-specific primer; Δ, primer specific for the targeted allele. (B) Southern blot of _Xho_I/_Hin_dIII-digested DNA from ES cells of the indicated genotypes. The probe is the 0.8-kb _Xho_I/_Eco_RI fragment shown in panel A. (C) PCR of wild-type and Terf1 _ex1_Δ/+ ES cells. (D) Northern blot of total RNA from Terf1+/+ and Terf1 _ex1_Δ/+ MEFs. Total RNA was hybridized first with radiolabeled Terf1 cDNA and subsequently with two oligonucleotides complementary to 7SK RNAs. “% Terf1 signal” refers to the signal derived from hybridization with the Terf1 cDNA normalized to loading (7SK signal). (E) Growth curves of Terf1+/+ and Terf1 _ex1_Δ/+ MEFs from E12.5 embryos. (F) Telomeric overhangs and telomeric restriction fragments from Terf1+/+ and Terf1 _ex1_Δ/+ MEFs obtained from littermates from two different litters. Molecular size markers are indicated in kilobase pairs. Relative G-strand overhang intensities were determined by normalization of the native (G-strand) signal to the signal obtained after denaturation (representing duplex TTAGGG repeats). The ratio of G-strand to duplex signals was arbitrarily set to 100 for the DNA in the first lane, and the other ratios were normalized to this value.
FIG. 2.
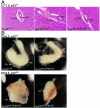
Morphologies of Terf1+/+, Terf1 _ex1_Δ/+, and Terf1 ex1_Δ/ex1_Δ embryos. (A) Hematoxylin-and-eosin-stained sections of E6 to E6.5 embryos (in utero) derived from intercrosses between Terf1 _ex1_Δ/+ mice. Extraembryonic (ex), embryonic (em), and ectoplacental-cone (epc) regions are indicated. Genotyping was performed by laser capture and PCR. (B) Hematoxylin-and-eosin-stained sections of E5 to E5.5 embryos (in utero) derived from intercrosses between Terf1 _ex1_Δ/+ mice. Note that the homozygous mutant embryos are grossly similar to their heterozygous and wild-type littermates.
FIG. 3.
p53 deficiency attenuates the embryonic lethality of the Terf1ex1Δ/ex1Δ genotype. (A) Hematoxylin-and-eosin-stained E7 to E7.5 sections of embryos (in utero) from intercrosses between _Terf1ex1Δ/+ p53_−/− mice. Extraembryonic (ex) and embryonic (em) tissues are indicated. Genotypes were identified by laser capture and PCR. Note that the Terf1ex1Δ/ex1Δ embryo was grossly normal, although smaller than its littermates. (B) E8 to E8.5 embryos derived from intercrosses between _Terf1ex1Δ/+ p53_−/− mice. Extraembryonic and embryonic regions are indicated.
FIG. 4.
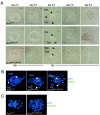
Terf1 ex1_Δ/ex1_Δ blastocysts show ICM cell death without detectable telomere fusions. (A) Day-3.5-postcoitus blastocysts isolated from Terf1 _ex1_Δ/+ intercrosses cultured in 96-well plates for 5 days. ICM and trophoblastic giant cells (TGC) are indicated. (B) TUNEL staining of Terf1+/+, Terf1 _ex1_Δ/+, and Terf1 ex1_Δ/ex1_Δ blastocysts. Blastocysts were cultured for 4 days and subjected to TUNEL staining. Green, TUNEL signal; blue, DNA stain (4′,6′-diamidino-2-phenylindole [DAPI]). (C) Telomeric FISH on metaphase spreads of Terf1+/+, Terf1 _ex1_Δ/+, and Terf1 ex1_Δ/ex1_Δ blastocysts at day 5.5. Cells were cultured for 2 days, and metaphase spreads were processed for telomeric FISH (green). DNA is stained with DAPI (blue).
Similar articles
- Telomere-associated protein TIN2 is essential for early embryonic development through a telomerase-independent pathway.
Chiang YJ, Kim SH, Tessarollo L, Campisi J, Hodes RJ. Chiang YJ, et al. Mol Cell Biol. 2004 Aug;24(15):6631-4. doi: 10.1128/MCB.24.15.6631-6634.2004. Mol Cell Biol. 2004. PMID: 15254230 Free PMC article. - POT1 as a terminal transducer of TRF1 telomere length control.
Loayza D, De Lange T. Loayza D, et al. Nature. 2003 Jun 26;423(6943):1013-8. doi: 10.1038/nature01688. Epub 2003 May 25. Nature. 2003. PMID: 12768206 - Role of Pin2/TRF1 in telomere maintenance and cell cycle control.
Zhou XZ, Perrem K, Lu KP. Zhou XZ, et al. J Cell Biochem. 2003 May 1;89(1):19-37. doi: 10.1002/jcb.10496. J Cell Biochem. 2003. PMID: 12682905 Review. - Regulation of telomerase by telomeric proteins.
Smogorzewska A, de Lange T. Smogorzewska A, et al. Annu Rev Biochem. 2004;73:177-208. doi: 10.1146/annurev.biochem.73.071403.160049. Annu Rev Biochem. 2004. PMID: 15189140 Review.
Cited by
- Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice.
Chiang YJ, Nguyen ML, Gurunathan S, Kaminker P, Tessarollo L, Campisi J, Hodes RJ. Chiang YJ, et al. Mol Cell Biol. 2006 Mar;26(6):2037-43. doi: 10.1128/MCB.26.6.2037-2043.2006. Mol Cell Biol. 2006. PMID: 16507984 Free PMC article. - TERRA regulate the transcriptional landscape of pluripotent cells through TRF1-dependent recruitment of PRC2.
Marión RM, Montero JJ, López de Silanes I, Graña-Castro O, Martínez P, Schoeftner S, Palacios-Fábrega JA, Blasco MA. Marión RM, et al. Elife. 2019 Aug 20;8:e44656. doi: 10.7554/eLife.44656. Elife. 2019. PMID: 31426913 Free PMC article. - TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis.
Muñoz P, Blanco R, de Carcer G, Schoeftner S, Benetti R, Flores JM, Malumbres M, Blasco MA. Muñoz P, et al. Mol Cell Biol. 2009 Mar;29(6):1608-25. doi: 10.1128/MCB.01339-08. Epub 2009 Jan 5. Mol Cell Biol. 2009. PMID: 19124610 Free PMC article. - Cyclin B-dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres.
McKerlie M, Zhu XD. McKerlie M, et al. Nat Commun. 2011 Jun 28;2:371. doi: 10.1038/ncomms1372. Nat Commun. 2011. PMID: 21712819 Free PMC article. - Telomerase reverse transcriptase-dependent telomere equilibration mitigates tissue dysfunction in mTert heterozygotes.
Meznikova M, Erdmann N, Allsopp R, Harrington LA. Meznikova M, et al. Dis Model Mech. 2009 Nov-Dec;2(11-12):620-6. doi: 10.1242/dmm.004069. Epub 2009 Oct 19. Dis Model Mech. 2009. PMID: 19841238 Free PMC article.
References
- Ancelin, K., M. Brunori, S. Bauwens, C. E. Koering, C. Brun, M. Ricoul, J. P. Pommier, L. Sabatier, and E. Gilson. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 22:3474-3487. - PMC - PubMed
- Artandi, S. E., S. Chang, S. L. Lee, S. Alson, G. J. Gottlieb, L. Chin, and R. A. DePinho. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406:641-645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM049046/GM/NIGMS NIH HHS/United States
- R37 GM049046/GM/NIGMS NIH HHS/United States
- R01 CA076027/CA/NCI NIH HHS/United States
- CA 76027/CA/NCI NIH HHS/United States
- GM 49046/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous