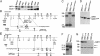Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice - PubMed (original) (raw)
Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice
Hwei-Ling Cheng et al. Proc Natl Acad Sci U S A. 2003.
Abstract
SIRT1 is a mammalian homolog of the Saccharomyces cerevisiae chromatin silencing factor Sir2. Dominant-negative and overexpression studies have implicated a role for SIRT1 in deacetylating the p53 tumor suppressor protein to dampen apoptotic and cellular senescence pathways. To elucidate SIRT1 function in normal cells, we used gene-targeted mutation to generate mice that express either a mutant SIRT1 protein that lacks part of the catalytic domain or has no detectable SIRT1 protein at all. Both types of SIRT1 mutant mice and cells had essentially the same phenotypes. SIRT1 mutant mice were small, and exhibited notable developmental defects of the retina and heart, and only infrequently survived postnatally. Moreover, SIRT1-deficient cells exhibited p53 hyperacetylation after DNA damage and increased ionizing radiation-induced thymocyte apoptosis. In SIRT1-deficient embryonic fibroblasts, however, p53 hyperacetylation after DNA damage was not accompanied by increased p21 protein induction or DNA damage sensitivity. Together, our observations provide direct evidence that endogenous SIRT1 protein regulates p53 acetylation and p53-dependent apoptosis, and show that the function of this enzyme is required for specific developmental processes.
Figures
Fig. 1.
Generation of SIRT1-deficient mice. (A) Western analysis of SIRT1 and tubulin expression in WT adult mouse tissues. (B) A schematic diagram of the germ-line SIRT1 locus, KOII targeting vector, and SIRT1Δex4 allele. SIRT1 exons comprising the conserved catalytic domain are dark gray. The relative locations of the 5′, 3′, and internal KOII probes are indicated. Restriction sites: BgI, _Bgl_I; N, _Not_I; S, _Sal_I; H, _Hin_dIII; E, _Eco_RI; Bg, _Bgl_II; B, _Bam_HI; Pac, _Pac_I; Swa, SwaI. (C) Southern blot of SIRT1+/Δex4, SIRT1+/NeoR, and SIRT1Δex4/Δex4 _Bgl_II-digested DNA with the internal KOII probe. (D) Western blot of SIRT1+/+, SIRT1+/Δex4, and SIRT1Δex4/Δex4 MEFs. SIRT1 and SIRT1Δex4 proteins are indicated. (E) A schematic diagram of the germ-line SIRT1 locus, SKO targeting vector, and SIRT1Δ allele, which are depicted as in A. (F) Southern blot of SIRT1+/Δneo and SIRT1+/Δ _Bgl_I/SwaI-digested DNA with the 5′SKO probe. (G) Western blot of SIRT1+/+, SIRT1+/Δ, and SIRT1Δ/Δ MEFs with anti-SIRT1 antibodies. Arrows indicate crossreacting bands that control for total protein.
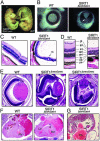
Fig. 2.
Developmental abnormalities in SIRT1-deficient mice. (A) WT and SIRT1Δex4/Δex4 E16.5 embryos. *, exencephaly in the SIRT1Δex4/Δex4 embryo. (B) Whole-mount eyes from E16.5 embryos. Arrow indicates open optic fissure in the SIRT1Δex4/Δex4 eye. (C) Retinal sections from 5-week-old mice. rs, rosette-like structure. (D) An enlargement of C. gl, ganglion cell layer; ipl, inner plexiform layer; inl, inner nuclear layer; opl, outer plexiform layer; onl, outer nuclear layer; is, inner segment; os, outer segment; pe, pigmented epithelium. (E) Retinal sections from E18.5 WT and SIRT1Δneo/Δneo embryos. (F) Sections through E18.5 hearts. *, the VSD. LV, left ventricle; RV, right ventricle. (G) Section through E18.5 SIRT1Δex4/Δex4 heart showing elongated atrioventricular valve (arrow).
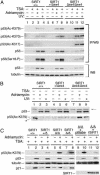
Fig. 3.
p53 hyperacetylation in SIRT1-deficient MEFs after DNA damage. (A) Western analysis of p53 IPs (IP/WB) and direct Western blots (WB) of whole-cell MEF lysates. Cells were incubated with adriamycin, were UV irradiated, and treated with 5 μM TSA as indicated. Blots were probed with antibodies specific for the indicated p53 modifications, total p53, and tubulin. (B) p53 hyperacetylation in the absence of TSA. Western blots of whole-cell extracts were probed as in A. *, a gel artifact in lane 1. Arrow indicates a crossreacting band that controls for total protein levels. (C) p53 acetylation in SIRT1+/+, SIRT1+/Δ, and two different SIRT1Δ/Δ MEF lines, and in SIRT1Δ/Δ MEFs infected with empty virus control (pBabe) or retroviral recombinant SIRT1 (rSIRT1).
Fig. 4.
Increased thymocyte apoptosis in SIRT1-deficient cells. (A) Western analysis of SIRT1 and SIRT1Δex4 protein in thymocytes of the indicated genotypes. (B) Western analysis of acetylated p53 in p53 IPs from SIRT1+/+ and SIRT1Δex4/Δex4 thymocytes, at 2.5 h after mock (–) or 500 rads IR (+), in the presence of 1 μM TSA. Western analyses on the IP inputs show total p53 levels, and the arrow indicates a crossreacting band that controls for total protein. (C) The percent nonapoptotic cells in SIRT1+/+, SIRT1+/Δex4, SIRT1Δex4/Δex4, and p53–/– CD4+ thymocytes after IR is shown.
Similar articles
- Role of NAD-dependent deacetylases SIRT1 and SIRT2 in radiation and cisplatin-induced cell death in vertebrate cells.
Matsushita N, Takami Y, Kimura M, Tachiiri S, Ishiai M, Nakayama T, Takata M. Matsushita N, et al. Genes Cells. 2005 Apr;10(4):321-32. doi: 10.1111/j.1365-2443.2005.00836.x. Genes Cells. 2005. PMID: 15773895 - Silent information regulator 2alpha, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes.
Alcendor RR, Kirshenbaum LA, Imai S, Vatner SF, Sadoshima J. Alcendor RR, et al. Circ Res. 2004 Nov 12;95(10):971-80. doi: 10.1161/01.RES.0000147557.75257.ff. Epub 2004 Oct 14. Circ Res. 2004. PMID: 15486319 - Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice.
Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Vakhrusheva O, et al. Circ Res. 2008 Mar 28;102(6):703-10. doi: 10.1161/CIRCRESAHA.107.164558. Epub 2008 Jan 31. Circ Res. 2008. PMID: 18239138 - Sirtuins and p53.
van Leeuwen I, Lain S. van Leeuwen I, et al. Adv Cancer Res. 2009;102:171-95. doi: 10.1016/S0065-230X(09)02005-3. Adv Cancer Res. 2009. PMID: 19595309 Review. - Retinal aging and sirtuins.
Ozawa Y, Kubota S, Narimatsu T, Yuki K, Koto T, Sasaki M, Tsubota K. Ozawa Y, et al. Ophthalmic Res. 2010;44(3):199-203. doi: 10.1159/000316484. Epub 2010 Sep 9. Ophthalmic Res. 2010. PMID: 20829644 Review.
Cited by
- Deciphering the potential role of post-translational modifications of histones in gastrointestinal cancers: a proteomics-based review with therapeutic challenges and opportunities.
Farrokhi Yekta R, Farahani M, Koushki M, Amiri-Dashatan N. Farrokhi Yekta R, et al. Front Oncol. 2024 Oct 21;14:1481426. doi: 10.3389/fonc.2024.1481426. eCollection 2024. Front Oncol. 2024. PMID: 39497715 Free PMC article. Review. - Melatonin mitigates doxorubicin induced chemo brain in a rat model in a NRF2/p53-SIRT1 dependent pathway.
Ebrahim NA, Elnagar MR, El-Gamal R, Habotta OA, Albadawi EA, Albadrani M, Bahashwan AS, Hassan HM. Ebrahim NA, et al. Heliyon. 2024 Sep 18;10(19):e38081. doi: 10.1016/j.heliyon.2024.e38081. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39386846 Free PMC article. - Sirt1: An Increasingly Interesting Molecule with a Potential Role in Bone Metabolism and Osteoporosis.
Chen Y, Xiao H, Liu Z, Teng F, Yang A, Geng B, Sheng X, Xia Y. Chen Y, et al. Biomolecules. 2024 Aug 8;14(8):970. doi: 10.3390/biom14080970. Biomolecules. 2024. PMID: 39199358 Free PMC article. Review. - A hypothesis: MiRNA-124 mediated regulation of sirtuin 1 and vitamin D receptor gene expression accelerates aging.
Dhar P, Moodithaya S, Patil P, Adithi K. Dhar P, et al. Aging Med (Milton). 2024 Jun 15;7(3):320-327. doi: 10.1002/agm2.12330. eCollection 2024 Jun. Aging Med (Milton). 2024. PMID: 38975301 Free PMC article. - The dual role of sirtuins in cancer: biological functions and implications.
Yu L, Li Y, Song S, Zhang Y, Wang Y, Wang H, Yang Z, Wang Y. Yu L, et al. Front Oncol. 2024 Jun 14;14:1384928. doi: 10.3389/fonc.2024.1384928. eCollection 2024. Front Oncol. 2024. PMID: 38947884 Free PMC article. Review.
References
- Imai, S., Armstrong, C. M., Kaeberlein, M. & Guarente, L. (2000) Nature 403, 795–800. - PubMed
- Denu, J. M. (2003) Trends Biochem. Sci. 28, 41–48. - PubMed
- Gasser, S. M. & Cockell, M. M. (2001) Gene 279, 1–16. - PubMed
- Guarente, L. (2000) Genes Dev. 14, 1021–1026. - PubMed
- Tsukamoto, Y., Kato, J. & Ikeda, H. (1997) Nature 388, 900–903. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous