Src phosphorylates Cas on tyrosine 253 to promote migration of transformed cells - PubMed (original) (raw)
Src phosphorylates Cas on tyrosine 253 to promote migration of transformed cells
Gary S Goldberg et al. J Biol Chem. 2003.
Abstract
Cas is a member of the focal adhesion complex. Phosphorylation of Cas by Src is an important event leading to cell transformation. Using mass spectrometry, we have mapped 11 sites in Cas that are phosphorylated by Src. These sites are all located between residues 132 and 414 of Cas, in a region that is required for binding to a number of other proteins including Crk. We tested synthetic peptides modeled on Cas phosphorylation sites, and found that the sequence containing tyrosine 253 was phosphorylated by Src most efficiently. Using cells derived from Cas-deficient mice, we confirmed that Cas greatly enhanced the ability of Src to transform cells. Phosphorylation of Cas on tyrosine 253 was not required for Src to increase growth rate, suppress contact inhibition, or suppress anchorage dependence. Yet, in contrast to these growth characteristics, phosphorylation of Cas on tyrosine 253 was required for Src to promote cell migration. Thus, a single phosphorylation site on this focal adhesion adaptor protein can effectively separate cell migration from other transformed growth characteristics.
Figures
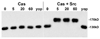
FIG. 1. Phosphorylation of Cas by Src
Full-length Cas was purified from baculovirus-infected Sf9 cells and incubated with ATP in the presence or absence of Src for 0, 20, or 60 min as indicated. After 60 min, one sample was incubated with Yersinia phosphatase (yop) for an additional 5 min. Samples were resolved by SDS-PAGE on 8% gels and examined by Western blotting with antibody directed against Cas. Phosphorylation of Cas was evident by a shift in migration from an apparent molecular mass of 130 kDa to about 170 kDa, which was reversed by phosphatase treatment.
FIG. 2. Mapping Src phosphorylation sites in Cas
Cas was incubated with ATP and Src or ATP alone as indicated, resolved by SDS-PAGE, and digested with trypsin. Resulting peptides were then examined by MALDI-TOF MS. Mass and intensity are shown on the x and y axis, respectively. Phosphorylated peptides, detected by an increase in mass of 80 daltons in the Src + Cas sample, are designated by the apparent phosphorylation sites they contain. The amino acid sequences of these fragments are listed in Table I.
FIG. 3. Phosphorylation of tyrosine 253 of Cas by Src
Cas was phosphorylated by Src, resolved by SDS-PAGE, digested with trypsin, and analyzed by LC-MS-MS. A deconvoluted chromatogram of the peptide containing residues 251–259 (251DIYDVPPVR259) is shown. Peaks belonging to the y-type ion series are indicated along the x axis, with their relative intensity shown on the y axis. The sequence is given above the chromatogram with the phosphorylation site indicated. The monoisotopic mass of each amino acid residue is given in parentheses below the sequence. Superscript 1, Val (97 daltons) + Pro (99 daltons) = 196 daltons; superscript 2, Tyr (163 daltons) + phosphate (80 daltons) = 243 daltons. These data indicate that tyrosine 253 of Cas was phosphorylated by Src.
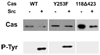
FIG. 4. Src phosphorylation sites lie between residues 118 and 423 in Cas
Wild type Cas (WT), CasY253F, and Cas118Δ423 were produced in bacteria and subjected to in vitro kinase assays with or without Src as indicated. Protein was then examined by Western blotting with anti-phosphotyrosine antibody (P-Tyr). Wild type Cas and CasY253F were phosphorylated by Src, but Cas118Δ423 was not, confirming that major Src phosphorylation sites were located between residues 118 and 423 in Cas.
FIG. 5. Diagram of Src phosphorylation sites and Cas transfection constructs
Tyrosine residues phosphorylated by Src are numbered over a schematic representation of Cas. Positions of the SH3 domain, proline-rich region (Pro), kinase substrate region, serine rich-region (Ser), Src binding sequence (SBS), and helix-loop-helix (HLH) motifs are indicated. Schematic diagrams of two mutants made for further testing are also shown. Tyrosine 253 was changed to phenylalanine to create CasY253F, and the entire substrate for the Src kinase region between residues 118 and 423 was deleted to create Cas118Δ423. WT, wild type.
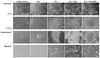
FIG. 6. Effects of Cas and Src on cell morphology, growth, and migration
Fibroblasts from homozygous null Cas knockout mice were transfected with wild type Cas or CasY253F in pBABEhygro, v-Src in pBABEpuro, or empty vectors and selected for resistance to puromycin and hygromycin. 20,000, 200, or 200,000 cells were plated on each well of 12-well tissue culture plates, 24-well low attachment plates, or Transwell inserts with 3.0-µm pores in 6-well cluster plates to examine anchored growth, nonanchored growth, or cell migration, respectively. Anchored cells grown for 2 days (subconfluent) and 6 days (confluent) are shown, along with nonanchored cells grown for 11 days and cells that migrated through a porous membrane to the bottom well of cluster plates over a 3-day period as indicated. Cas was not required for Src to cause transformation of cell morphology but was required for Src-transformed cells to achieve robust anchorage independence at these plating densities. Phosphorylation of Cas at tyrosine 253 was required for Src to induce cell migration, but not anchorage independence. (Bar = 250 µm)
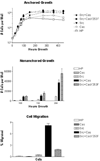
FIG. 7. Phosphorylation of tyrosine 253 is important for cell migration, but not for anchored or nonanchored growth
Cells were plated as described in the legend for Fig. 6 and counted at the indicated time points to examine anchored growth or nonanchored growth. To measure cell migration, the percent of cells that migrated through the membranes to the bottom well after 72 h was calculated as shown. Data are shown as mean ± S.E. with n = 2, 4, and 3 for anchored growth, nonanchored growth, and cell migration, respectively. Phosphorylation of Cas at tyrosine 253 was required for Src to induce cell migration but not to reduce contact growth inhibition or achieve anchorage independence. HP, empty hygro/puro vectors.
FIG. 8. Src activity is independent of Cas
Cells were examined by Western blotting for v-Src, active Src, and β-actin as indicated. Cas and Src were expressed in the appropriate transfectants. Src activity wasnot affected by Cas expression.
FIG. 9. Phosphorylation of Cas at tyrosine 253 is not required for Crk binding
Crk was immunoprecipitated (IP) from cells transfected with Src and wild type Cas (WT) or Src and CasY253F. Immunoprecipitated protein was then analyzed by Western blotting for Cas and Crk as indicated. Both wild type Cas and CasY253F coprecipitated with Crk.
Similar articles
- SRC uses Cas to suppress Fhl1 in order to promote nonanchored growth and migration of tumor cells.
Shen Y, Jia Z, Nagele RG, Ichikawa H, Goldberg GS. Shen Y, et al. Cancer Res. 2006 Feb 1;66(3):1543-52. doi: 10.1158/0008-5472.CAN-05-3152. Cancer Res. 2006. PMID: 16452211 - Crk-associated substrate tyrosine phosphorylation sites are critical for invasion and metastasis of SRC-transformed cells.
Brábek J, Constancio SS, Siesser PF, Shin NY, Pozzi A, Hanks SK. Brábek J, et al. Mol Cancer Res. 2005 Jun;3(6):307-15. doi: 10.1158/1541-7786.MCR-05-0015. Mol Cancer Res. 2005. PMID: 15972849 - Differential effect of the focal adhesion kinase Y397F mutant on v-Src-stimulated cell invasion and tumor growth.
Chang LC, Huang CH, Cheng CH, Chen BH, Chen HC. Chang LC, et al. J Biomed Sci. 2005;12(4):571-85. doi: 10.1007/s11373-005-7212-5. Epub 2005 Nov 10. J Biomed Sci. 2005. PMID: 16132110 - Tyrosine phosphorylation within the SH3 domain regulates CAS subcellular localization, cell migration, and invasiveness.
Janoštiak R, Tolde O, Brůhová Z, Novotný M, Hanks SK, Rösel D, Brábek J. Janoštiak R, et al. Mol Biol Cell. 2011 Nov;22(22):4256-67. doi: 10.1091/mbc.E11-03-0207. Epub 2011 Sep 21. Mol Biol Cell. 2011. PMID: 21937722 Free PMC article. - The role of SRC-CAS interactions in cellular transformation: ectopic expression of the carboxy terminus of CAS inhibits SRC-CAS interaction but has no effect on cellular transformation.
Burnham MR, Harte MT, Bouton AH. Burnham MR, et al. Mol Carcinog. 1999 Sep;26(1):20-31. doi: 10.1002/(sici)1098-2744(199909)26:1<20::aid-mc3>3.0.co;2-m. Mol Carcinog. 1999. PMID: 10487518
Cited by
- The role of Src kinase in macrophage-mediated inflammatory responses.
Byeon SE, Yi YS, Oh J, Yoo BC, Hong S, Cho JY. Byeon SE, et al. Mediators Inflamm. 2012;2012:512926. doi: 10.1155/2012/512926. Epub 2012 Nov 11. Mediators Inflamm. 2012. PMID: 23209344 Free PMC article. Review. - Processive phosphorylation: mechanism and biological importance.
Patwardhan P, Miller WT. Patwardhan P, et al. Cell Signal. 2007 Nov;19(11):2218-26. doi: 10.1016/j.cellsig.2007.06.006. Epub 2007 Jun 22. Cell Signal. 2007. PMID: 17644338 Free PMC article. Review. - Independent effects of Src kinase and podoplanin on anchorage independent cell growth and migration.
Retzbach EP, Sheehan SA, Krishnan H, Zheng H, Zhao C, Goldberg GS. Retzbach EP, et al. Mol Carcinog. 2022 Jul;61(7):677-689. doi: 10.1002/mc.23410. Epub 2022 Apr 26. Mol Carcinog. 2022. PMID: 35472679 Free PMC article. - Defining the substrate specificity determinants recognized by the active site of C-terminal Src kinase-homologous kinase (CHK) and identification of β-synuclein as a potential CHK physiological substrate.
Ia KK, Jeschke GR, Deng Y, Kamaruddin MA, Williamson NA, Scanlon DB, Culvenor JG, Hossain MI, Purcell AW, Liu S, Zhu HJ, Catimel B, Turk BE, Cheng HC. Ia KK, et al. Biochemistry. 2011 Aug 9;50(31):6667-77. doi: 10.1021/bi2001938. Epub 2011 Jul 18. Biochemistry. 2011. PMID: 21699177 Free PMC article. - Cooperative activation of Src family kinases by SH3 and SH2 ligands.
Yadav SS, Miller WT. Yadav SS, et al. Cancer Lett. 2007 Nov 8;257(1):116-23. doi: 10.1016/j.canlet.2007.07.012. Epub 2007 Aug 24. Cancer Lett. 2007. PMID: 17719722 Free PMC article.
References
- Frisch SM, Screaton RA. Curr. Opin. Cell Biol. 2001;13:555–562. - PubMed
- Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. - PubMed
- Schlaepfer DD, Hauck CR, Sieg DJ. Prog. Biophys. Mol. Biol. 1999;71:435–478. - PubMed
- Sieg DJ, Hauck CR, Schlaepfer DD. J. Cell Sci. 1999;112:2677–2691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA 58530/CA/NCI NIH HHS/United States
- R01 CA088805/CA/NCI NIH HHS/United States
- R01 CA058530/CA/NCI NIH HHS/United States
- CA 88805-01A2/CA/NCI NIH HHS/United States
- R01 CA058530-10A1/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous