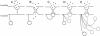Positive selection on protein-length in the evolution of a primate sperm ion channel - PubMed (original) (raw)
Comparative Study
. 2003 Oct 14;100(21):12241-6.
doi: 10.1073/pnas.2033555100. Epub 2003 Oct 1.
Affiliations
- PMID: 14523237
- PMCID: PMC218743
- DOI: 10.1073/pnas.2033555100
Comparative Study
Positive selection on protein-length in the evolution of a primate sperm ion channel
Ondrej Podlaha et al. Proc Natl Acad Sci U S A. 2003.
Abstract
Positive Darwinian selection on advantageous point substitutions has been demonstrated in many genes. We here provide empirical evidence, for the first time, that positive selection can also act on insertion/deletion (indel) substitutions in the evolution of a protein. CATSPER1 is a voltage-gated calcium channel found exclusively in the plasma membrane of the mammalian sperm tail and it is essential for sperm motility. We determined the DNA sequences of the first exon of the CATSPER1 gene from 15 primates, which encodes the intracellular N terminus region of approximately equal to 400 aa. These sequences exhibit an excessively high frequency of indels. However, all indels have lengths that are multiples of 3 nt (3n indels) and do not disrupt the ORF. The number of indel substitutions per site per year in CATSPER1 is five to eight times the corresponding rates calculated from two large-scale primate genomic comparisons, which represent the neutral rate of indel substitutions. Moreover, CATSPER1 indels are considerably longer than neutral indels. These observations strongly suggest that positive selection has been promoting the fixation of indel mutations in CATSPER1 exon 1. It has been shown in certain ion channels that the length of the N terminus region affects the rate of channel inactivation. This finding suggests that the selection detected may be related to the regulation of the CATSPER1 channel, which can affect sperm motility, an important determinant in sperm competition.
Figures
Fig. 1.
Amino acid sequence alignment of the exon 1 of CATSPER1 from 16 primates. This alignment is derived from
clustal x
with default parameters. The dots represent an amino acid identical to the first sequence, and dashes represent an alignment gap. Asterisks show five sites at which positive selection for amino acid substitutions is inferred by the likelihood method with >95% posterior probabilities (see text).
Fig. 2.
Phylogeny of the 16 primates studied here. The numbers on tree branches are parsimony-inferred numbers of indel substitutions in CATSPER1 exon 1, based on the alignment shown in Fig. 1.
Fig. 3.
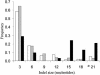
Size distribution of 3_n_ indels. White and gray bars represent 3_n_ indels of the genomic data from refs. and , respectively, and black bars represent 3_n_ indels from CATSPER1 exon 1 sequences of 16 primates. The frequencies are calculated by the number of indels of a particular size divided by the total number of 3_n_ indels.
Fig. 4.
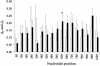
Sliding-window analysis of CATSPER1 exon 1 sequences from 15 primates. The lemur sequence was not used here because of the presence of a large number of indels. The nonoverlapping window size is 20 codons. Average numbers of synonymous (_d_S) and nonsynonymous (_d_N) substitutions per site among the 15 sequences are shown by open and filled bars, respectively, with the error bars representing one standard error. One window has a marginally significant _d_N > _d_S (Z test, P = 0.053), and it is indicated by an asterisk. The nucleotide positions are from an alignment of the 15 sequences with all of the indels removed and thus do not directly correspond to the positions in Fig. 1.
Fig. 5.
Schematics of the “ball-and-chain” model of channel inactivation. (A) Closed state. (B) Open state. (C) Inactivated state. According to this model, the N terminus represents a tethered plug that can physically block the intracellular side of the ion channel pore and cause inactivation. Different lengths of the N terminus region result in different rates of channel inactivation (45, 46), where a shorter N terminus (D) causes a more rapid inactivation in comparison to a longer N terminus (E).
Fig. 6.
The evolutionary relationship of CATSPER1 with mammalian Kv, Nav, and Cav channels. The tree was reconstructed with the N terminus region of each ion channel, which was determined by hydropathy analysis. The neighbor-joining method with protein p distance (29) was used. Numbers at interior nodes are bootstrap percentages from 1,000 replications. Branch lengths are drawn to scale (number of amino acid substitutions per site). The root of the tree was determined to be on the deepest branch shown here, by using Drosophila and vertebrate inward-rectifier K channels as outgroups (49). When the entire sequences of the ion channels are used, the tree topology remains the same with the exception of the interrelationships among KV1.4, KV3.3, and KV4.3. The GenBank accession numbers are: human KV1.4, A39922; mouse KV1.4, NP_067250.1; human KV3.3, NP_004968.1; mouse KV3.3, Q63959; human KV4.3, NP_004971.1; mouse KV4.3, NP_064315.1; human CaV2.2, Q00975; mouse CaV2.2, O55017; human CaV1.2, NP_000710.1; mouse CaV1.2, A44467; human CaV1.1, A55645; mouse CaV1.1, NP_055008.1; human NaV1.3, Q9NY46; mouse NaV1.3, NP_035453.1; human NaV1.5, Q14524; mouse NaV1.5, NP_067519.1; human CATSPER1, AF407333_1; mouse Catsper1, AF407332_1.
Similar articles
- Positive selection for indel substitutions in the rodent sperm protein catsper1.
Podlaha O, Webb DM, Tucker PK, Zhang J. Podlaha O, et al. Mol Biol Evol. 2005 Sep;22(9):1845-52. doi: 10.1093/molbev/msi178. Epub 2005 Jun 1. Mol Biol Evol. 2005. PMID: 15930155 Free PMC article. - Structural evolution of CatSper1 in rodents is influenced by sperm competition, with effects on sperm swimming velocity.
Vicens A, Tourmente M, Roldan ER. Vicens A, et al. BMC Evol Biol. 2014 May 16;14:106. doi: 10.1186/1471-2148-14-106. BMC Evol Biol. 2014. PMID: 24884901 Free PMC article. - Degeneration of an intracellular ion channel in the primate lineage by relaxation of selective constraints.
Cai X, Patel S. Cai X, et al. Mol Biol Evol. 2010 Oct;27(10):2352-9. doi: 10.1093/molbev/msq122. Epub 2010 May 12. Mol Biol Evol. 2010. PMID: 20463046 - Ion channels in sperm physiology and male fertility and infertility.
Shukla KK, Mahdi AA, Rajender S. Shukla KK, et al. J Androl. 2012 Sep-Oct;33(5):777-88. doi: 10.2164/jandrol.111.015552. Epub 2012 Mar 22. J Androl. 2012. PMID: 22441763 Review. - CatSper channel, sperm function and male fertility.
Singh AP, Rajender S. Singh AP, et al. Reprod Biomed Online. 2015 Jan;30(1):28-38. doi: 10.1016/j.rbmo.2014.09.014. Epub 2014 Oct 8. Reprod Biomed Online. 2015. PMID: 25457194 Review.
Cited by
- Intrinsically disordered proteins: Ensembles at the limits of Anfinsen's dogma.
Kulkarni P, Leite VBP, Roy S, Bhattacharyya S, Mohanty A, Achuthan S, Singh D, Appadurai R, Rangarajan G, Weninger K, Orban J, Srivastava A, Jolly MK, Onuchic JN, Uversky VN, Salgia R. Kulkarni P, et al. Biophys Rev (Melville). 2022 Mar 17;3(1):011306. doi: 10.1063/5.0080512. eCollection 2022 Mar. Biophys Rev (Melville). 2022. PMID: 38505224 Free PMC article. Review. - Unconventional conservation reveals structure-function relationships in the synaptonemal complex.
Kursel LE, Cope HD, Rog O. Kursel LE, et al. Elife. 2021 Nov 17;10:e72061. doi: 10.7554/eLife.72061. Elife. 2021. PMID: 34787570 Free PMC article. - Comparative genomic analysis suggests that the sperm-specific sodium/proton exchanger and soluble adenylyl cyclase are key regulators of CatSper among the Metazoa.
Romero F, Nishigaki T. Romero F, et al. Zoological Lett. 2019 Jul 26;5:25. doi: 10.1186/s40851-019-0141-3. eCollection 2019. Zoological Lett. 2019. PMID: 31372239 Free PMC article. - Transition-transversion mutations in the polyketide synthase gene of Aspergillus section Nigri.
Thomas BT, Ogunkanmi LA, Iwalokun BA, Popoola OD. Thomas BT, et al. Heliyon. 2019 Jun 15;5(6):e01881. doi: 10.1016/j.heliyon.2019.e01881. eCollection 2019 Jun. Heliyon. 2019. PMID: 31338447 Free PMC article. - Spontaneous Switching among Conformational Ensembles in Intrinsically Disordered Proteins.
Choi UB, Sanabria H, Smirnova T, Bowen ME, Weninger KR. Choi UB, et al. Biomolecules. 2019 Mar 22;9(3):114. doi: 10.3390/biom9030114. Biomolecules. 2019. PMID: 30909517 Free PMC article. Review.
References
- Hughes, A. L. (1999) Adaptive Evolution of Genes and Genomes (Oxford Univ. Press, New York).
- Wolfe, K. H. & Li, W. H. (2003) Nat. Genet. 33, Suppl., 255–265. - PubMed
- Hughes, A. L. & Nei, M. (1988) Nature 335, 167–170. - PubMed
- Tanaka, T. & Nei, M. (1989) Mol. Biol. Evol. 6, 447–459. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials