Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP - PubMed (original) (raw)
Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP
Zhong Deng et al. J Virol. 2003 Nov.
Abstract
Epstein-Barr virus OriP confers cell cycle-dependent DNA replication and stable maintenance on plasmids in EBNA1-positive cells. The dyad symmetry region of OriP contains four EBNA1 binding sites that are punctuated by 9-bp repeats referred to as nonamers. Previous work has shown that the nonamers bind to cellular factors associated with human telomeres and contribute to episomal maintenance of OriP. In this work, we show that substitution mutation of all three nonamer sites reduces both DNA replication and plasmid maintenance of OriP-containing plasmids by 2.5- to 5-fold. The nonamers were required for high-affinity binding of TRF1, TRF2, and hRap1 to the dyad symmetry element but were not essential for the binding of EBNA1 as determined by DNA affinity purification from nuclear extracts. Chromatin immunoprecipitation assays indicated that TRF1, TRF2, and hRap1 bound OriP in vivo. Cell cycle studies indicate that TRF2 binding to OriP peaks in G(1)/S while TRF1 binding peaks in G(2)/M. OriP replication was inhibited by transfection of full-length TRF1 but not by deletion mutants lacking the myb DNA binding domain. In contrast, OriP replication was not affected by transfection of full-length TRF2 or hRap1 but was potently inhibited by dominant-negative TRF2 or hRap1 amino-terminal truncation mutants. Knockdown experiments with short interfering RNAs (siRNAs) directed against TRF2 and hRap1 severely reduced OriP replication, while TRF1 siRNA had a modest stimulatory effect on OriP replication. These results indicate that TRF2 and hRap1 promote, while TRF1 antagonizes, OriP-dependent DNA replication and suggest that these telomeric factors contribute to the establishment of replication competence at OriP.
Figures
FIG. 1.
Nonamer repeats contribute to DNA replication and plasmid maintenance of OriP. (A) Sequences of OriPwt and OriPnm− with substitution mutations indicated by underlining. Black spheres indicate EBNA1 binding sites. Gray spheres indicate TRF2 and/or TRF1 binding sites. (B) HeLa, 293, and D98/HR1 cells were transfected with OriPwt or OriPnm− plasmid and assayed for DNA replication at 24 or 72 h posttransfection. _Bam_HI digests are shown in the top panel and _Dpn_I/_Bam_HI double digests are indicated in the bottom panel for each cell type indicated to the left. (C) Raji cells were electroporated with OriPwt or OriPnm− and pHEBO control. Equal numbers of GFP-positive cells were selected by FACS and then assayed at 72 h posttransfection for DNA replication based on _Dpn_I resistance. _Dpn_I/_Bam_HI digests (top panel) and _Bam_HI digests (lower panel) were detected by Southern blotting and quantitated by PhosphorImager analysis. (D) Raji cells were electroporated and selected by FACS as described for panel A. After 21 days in culture without selection, DNA was extracted and analyzed by Southern blotting after _Bam_HI digestion (top panel) or was uncut (bottom panel). Quantitation of Southern blots by PhosphorImager analysis is indicated below.
FIG. 2.
Telomere repeat binding factors interact with nonamer repeats in Raji cell nuclear extracts. Raji whole-cell extracts were subjected to DNA affinity purification with biotinylated DNA derived from pBKS, DSwt, DSnm−, or OriPΔDS. Affinity-purified proteins were washed under low (A)- or high (B)-stringency conditions and assayed by Western blotting with antibodies specific for EBNA1, TRF2, TRF1, hRap1, PCNA, and acetyl-histone H3.
FIG. 3.
TRF1, TRF2, and hRap1 bind OriP in vivo. (A) Raji cells were formaldehyde cross-linked and subjected to ChIP assay with antibodies to EBNA1, TRF2, hRap1, TRF1, or control antisera to monoclonal Flag or from preimmune rabbit serum. Immunoprecipitated DNA was amplified with primers specific for OriP or the BZLF1 promoter region. (B) D98/HR1 cells were transfected with OriPwt or OriPnm− cells, sorted for GFP-positive cells, and assayed by ChIP 72 h posttransfection. Primers specific for plasmid-based OriP (OriPwt and OriPnm−) or the ampicillin gene (Amp) were used to amplify immunoprecipitated DNA. Quantification of several experiments is indicated in the bar graph to the right.
FIG. 4.
Cell cycle-dependent binding of TRF1 to OriP. (A) Raji cells were arrested in G0 by serum starvation and assayed by ChIP for association of EBNA1, TRF2, or TRF1 with OriP or control BMRF1 promoter DNA. (B) Raji cells were synchronized by double thymidine block and isolated at various stages of the cell cycle. Asynchronous (A), G1, S, and G2/M phase-arrested cells were assayed by ChIP for EBNA1, TRF2, and TRF1 binding to OriP. (C) FACS profile of Raji cells synchronized in panel B as described above. Raji cells were stained with propidium iodide prior to analysis by FACS. IgG, immunoglobulin G.
FIG. 5.
Truncation mutants of telomere repeat factors inhibit OriP replication. (A) OriPwt or control plasmid ΔOriP was transfected into ZKO-293 cells with pCMV-Flag-2 vector or an expression plasmid for hRap1, TRF1, or TRF2 and their derivatives as indicated above each lane. Replicated DNA was measured by _Dpn_I resistance in the top panel (_Dpn_I/_Bam_HI), and total extracted DNA is indicated in the bottom panel (_Bam_HI digest). (B) PhosphorImager quantitation of at least three independent Southern blots is presented as a bar graph. (C) Transfected proteins were detected by Western blotting with anti-Flag antibody (full-length hRap1 is not Flag tagged), or the same blot was reprobed with anti-TRF2 (full-length TRF2 is not Flag tagged). (D) Schematic diagram of truncation mutants cloned into pCMV-Flag-2. The hRap1 BRCT (BRCA1 C-terminal homology), myb DNA binding, TRF2 interaction (TRF2-IX), and TRF homodimerization (TRFH) domains are indicated.
FIG. 6.
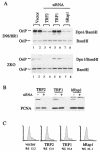
siRNA knockdown mutants of TRF2 and hRap1 inhibit OriP replication. (A) D98/HR1 cells were cotransfected with OriPwt and siRNA expression plasmids for TRF2, TRF1, and hRap1. _Dpn_I-resistant replicated DNA is indicated in the top panel (_Dpn_I/_Bam_HI), and total extracted DNA is indicated in the bottom panel (_Bam_HI). (B) Western blots of cell extracts derived from siRNA-transfected cells (+) or vector-transfected cells (−) were probed with antibody specific to the siRNA target indicated above (top panel) or with antibody to PCNA (bottom panel). (C) FACS profile of propidium iodide-treated cells that were transfected with vector, TRF2, TRF1, or hRap1 siRNA plasmids, as indicated. The percentage of cells in S phase is indicated below.
Similar articles
- ORC binding to TRF2 stimulates OriP replication.
Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. Atanasiu C, et al. EMBO Rep. 2006 Jul;7(7):716-21. doi: 10.1038/sj.embor.7400730. Epub 2006 Jun 16. EMBO Rep. 2006. PMID: 16799465 Free PMC article. - Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication.
Deng Z, Lezina L, Chen CJ, Shtivelband S, So W, Lieberman PM. Deng Z, et al. Mol Cell. 2002 Mar;9(3):493-503. doi: 10.1016/s1097-2765(02)00476-8. Mol Cell. 2002. PMID: 11931758 - Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats.
Ohki R, Ishikawa F. Ohki R, et al. Nucleic Acids Res. 2004 Mar 8;32(5):1627-37. doi: 10.1093/nar/gkh309. Print 2004. Nucleic Acids Res. 2004. PMID: 15007108 Free PMC article. - Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance.
Walker JR, Zhu XD. Walker JR, et al. Mech Ageing Dev. 2012 Jun;133(6):421-34. doi: 10.1016/j.mad.2012.05.002. Epub 2012 May 23. Mech Ageing Dev. 2012. PMID: 22634377 Review. - EBNA1 and host factors in Epstein-Barr virus latent DNA replication.
Frappier L. Frappier L. Curr Opin Virol. 2012 Dec;2(6):733-9. doi: 10.1016/j.coviro.2012.09.005. Epub 2012 Sep 30. Curr Opin Virol. 2012. PMID: 23031715 Review.
Cited by
- A protein array screen for Kaposi's sarcoma-associated herpesvirus LANA interactors links LANA to TIP60, PP2A activity, and telomere shortening.
Shamay M, Liu J, Li R, Liao G, Shen L, Greenway M, Hu S, Zhu J, Xie Z, Ambinder RF, Qian J, Zhu H, Hayward SD. Shamay M, et al. J Virol. 2012 May;86(9):5179-91. doi: 10.1128/JVI.00169-12. Epub 2012 Feb 29. J Virol. 2012. PMID: 22379092 Free PMC article. - EBV latency types adopt alternative chromatin conformations.
Tempera I, Klichinsky M, Lieberman PM. Tempera I, et al. PLoS Pathog. 2011 Jul;7(7):e1002180. doi: 10.1371/journal.ppat.1002180. Epub 2011 Jul 28. PLoS Pathog. 2011. PMID: 21829357 Free PMC article. - Epstein-Barr nuclear antigen 1 (EBNA1)-dependent recruitment of origin recognition complex (Orc) on oriP of Epstein-Barr virus with purified proteins: stimulation by Cdc6 through its direct interaction with EBNA1.
Moriyama K, Yoshizawa-Sugata N, Obuse C, Tsurimoto T, Masai H. Moriyama K, et al. J Biol Chem. 2012 Jul 6;287(28):23977-94. doi: 10.1074/jbc.M112.368456. Epub 2012 May 14. J Biol Chem. 2012. PMID: 22589552 Free PMC article. - ORC binding to TRF2 stimulates OriP replication.
Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. Atanasiu C, et al. EMBO Rep. 2006 Jul;7(7):716-21. doi: 10.1038/sj.embor.7400730. Epub 2006 Jun 16. EMBO Rep. 2006. PMID: 16799465 Free PMC article. - Trf1 is not required for proliferation or functional telomere maintenance in chicken DT40 cells.
Cooley C, Baird KM, Faure V, Wenner T, Stewart JL, Modino S, Slijepcevic P, Farr CJ, Morrison CG. Cooley C, et al. Mol Biol Cell. 2009 May;20(10):2563-71. doi: 10.1091/mbc.e08-10-1019. Epub 2009 Mar 25. Mol Biol Cell. 2009. PMID: 19321665 Free PMC article.
References
- Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous