Chromatin acetylation and remodeling at the Cis promoter during STAT5-induced transcription - PubMed (original) (raw)
Chromatin acetylation and remodeling at the Cis promoter during STAT5-induced transcription
Anne Rascle et al. Nucleic Acids Res. 2003.
Abstract
The signal transducer and activator of transcription STAT5 plays a major role in cytokine-induced expression of genes involved in cell proliferation and survival. Although several STAT5 partners have been identified, the molecular events taking place at the promoter level upon STAT5 recruitment have not yet been characterized in great detail. Using chromatin immunoprecipitation and accessibility assays, we characterized histone acetylation and chromatin remodeling events occurring during transcriptional activation of the endogenous murine Cis gene, a STAT5 target gene, in response to IL-3. We found that STAT5 binding in vivo is associated with low histone H3 and H4 acetylation levels in the proximity of the STAT5 binding sites. STAT5 recruitment also results in chromatin reorganization over that promoter region. These events (STAT5 binding, histone acetylation and chromatin remodeling) are not sufficient for transcriptional activation, which requires a non-histone protein deacetylase. These data reveal novel implications of STAT5 in chromatin regulation during cytokine-induced transcription, thus contributing to a better understanding of the mechanism of transcriptional activation by STAT5.
Figures
Figure 1
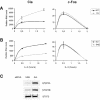
STAT5-dependent and -independent expression of Cis and c-Fos in Ba/F3 cells stimulated with IL-3. (A) Cis but not c-Fos is up-regulated in cells expressing a constitutively active form of STAT5A. Ba/F3-WT and Ba/F3-1*6 cells were stimulated with IL-3, and mRNAs levels for Cis and c-Fos analyzed by real-time PCR, as described in Materials and Methods. (B) Cis but not c-Fos induction is abrogated in STAT5 knock-down cells. Ba/F3-β cells were transfected with siRNA duplexes specific for both STAT5A and STAT5B isoforms (5AB) or with a non-specific control (ScI), as described in Materials and Methods. Thirty-one hours post-transfection, cells were stimulated with IL-3 and mRNAs levels analyzed as above. (C) STAT5 protein levels were monitored by western blot from STAT5 knock-down (5AB) and control (ScI) cells 31 h following siRNA transfection. As a control for specificity, STAT3 expression was shown to be unaffected following STAT5 knock-down at both the protein (this figure) and RNA (not shown) level.
Figure 2
Schematic representation of murine Cis (A) and c-Fos (B) genes. Amplicons analyzed by ChIP are indicated as horizontal double-headed arrows. Putative transcription factor binding sites are represented as boxes. Positions are relative to the transcription start (or CAP) site (+1). ORF, open reading frame. Cis gene contains no intron and c-Fos gene three introns.
Figure 3
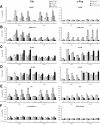
Regulation of histone H3 and H4 acetylation along the Cis and c-Fos genes upon IL-3 stimulation. Ba/F3-β cells were stimulated with IL-3 in the absence or presence of TSA, and ChIP assays performed as described in Materials and Methods, using antibodies specific for STAT5A+B (STAT5) (A), RNA polymerase II (RNA Pol II) (B), acetylated histone H3 (Ac-H3) (C), acetylated histone H4 (Ac-H4) (D), non-acetylated histone H4 (H4) (E) or no antibody as a control (F). Amplicons indicated in Figure 2 specific for Cis and c-Fos genes were analyzed by real-time PCR (Materials and Methods).
Figure 4
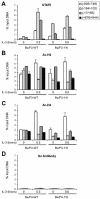
STAT5 is constitutively bound to the Cis promoter in cells expressing STAT5A-1*6, a constitutively active form of STAT5A. Unstimulated and IL-3-stimulated (30 min) Ba/F3-WT and Ba/F3-1*6 cells were analyzed by ChIP, using antibodies specific for STAT5A+B (STAT5) (A), acetylated histone H3 (Ac-H3) (B), acetylated histone H4 (Ac-H4) (C) or no antibody as a control (D), as described above. Amplicons specific for Cis were analyzed by real-time PCR as before.
Figure 5
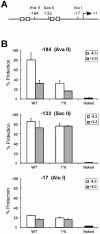
Chromatin is constitutively accessible to restriction enzyme digestion at position –184 of the Cis promoter in cells expressing STAT5A-1*6. (A) Structure of the Cis promoter, indicating the position of the restriction sites analyzed by CHART-PCR. Gray boxes represent STAT5 binding sites. (B) Nuclei from unstimulated and IL-3-stimulated (30 min) Ba/F3-WT and Ba/F3-1*6 cells were analyzed by CHART-PCR, as described in Materials and Methods. Purified genomic (naked) DNA was used as to control the cutting efficiency of the restriction enzyme.
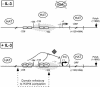
Figure 6
Model of STAT5-regulated transcription of Cis in response to IL-3. In unstimulated Ba/F3 cells, the level of histone H3 and H4 acetylation along the Cis gene is high, probably through the action of HAT complexes. The absence of effect of TSA suggests that the gene is not targeted by HDACs. Upon IL-3 stimulation, STAT5 (ST5) is recruited to its binding sites (gray boxes). STAT5 binding results in a local decrease in histone H3/H4 acetylation, and in chromatin remodeling at position –184, possibly through nucleosome sliding (dashed oval). STAT5 also allows, directly or indirectly, an as yet uncharacterized TSA-sensitive factor (gray diamond) to participate in the recruitment of the basal transcription machinery (PIC, pre-initiation complex), in turn permitting transcriptional initiation to occur (22).
Similar articles
- Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5.
Rascle A, Johnston JA, Amati B. Rascle A, et al. Mol Cell Biol. 2003 Jun;23(12):4162-73. doi: 10.1128/MCB.23.12.4162-4173.2003. Mol Cell Biol. 2003. PMID: 12773560 Free PMC article. - Transcriptional regulation of the beta-casein gene by cytokines: cross-talk between STAT5 and other signaling molecules.
Chida D, Wakao H, Yoshimura A, Miyajima A. Chida D, et al. Mol Endocrinol. 1998 Nov;12(11):1792-806. doi: 10.1210/mend.12.11.0196. Mol Endocrinol. 1998. PMID: 9817603 - CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation.
Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. Matsumoto A, et al. Blood. 1997 May 1;89(9):3148-54. Blood. 1997. PMID: 9129017 - Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer.
Wingelhofer B, Neubauer HA, Valent P, Han X, Constantinescu SN, Gunning PT, Müller M, Moriggl R. Wingelhofer B, et al. Leukemia. 2018 Aug;32(8):1713-1726. doi: 10.1038/s41375-018-0117-x. Epub 2018 Mar 27. Leukemia. 2018. PMID: 29728695 Free PMC article. Review. - Chromatin remodeling and transcription.
Hirose S. Hirose S. J Biochem. 1998 Dec 1;124(6):1060-4. doi: 10.1093/oxfordjournals.jbchem.a022220. J Biochem. 1998. PMID: 9832607 Review.
Cited by
- Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer.
Pinz S, Unser S, Rascle A. Pinz S, et al. BMC Mol Biol. 2016 Apr 14;17:10. doi: 10.1186/s12867-016-0063-y. BMC Mol Biol. 2016. PMID: 27074708 Free PMC article. - Erythropoietin Regulates Transcription and YY1 Dynamics in a Pre-established Chromatin Architecture.
Perreault AA, Brown JD, Venters BJ. Perreault AA, et al. iScience. 2020 Sep 20;23(10):101583. doi: 10.1016/j.isci.2020.101583. eCollection 2020 Oct 23. iScience. 2020. PMID: 33089097 Free PMC article. - p65 Negatively regulates transcription of the cyclin E gene.
Janbandhu VC, Singh AK, Mukherji A, Kumar V. Janbandhu VC, et al. J Biol Chem. 2010 Jun 4;285(23):17453-64. doi: 10.1074/jbc.M109.058974. Epub 2010 Apr 11. J Biol Chem. 2010. PMID: 20385564 Free PMC article. - Identification of human STAT5-dependent gene regulatory elements based on interspecies homology.
Nelson EA, Walker SR, Li W, Liu XS, Frank DA. Nelson EA, et al. J Biol Chem. 2006 Sep 8;281(36):26216-24. doi: 10.1074/jbc.M605001200. Epub 2006 Jul 13. J Biol Chem. 2006. PMID: 16840779 Free PMC article. - The natural chemopreventive agent sulforaphane inhibits STAT5 activity.
Pinz S, Unser S, Rascle A. Pinz S, et al. PLoS One. 2014 Jun 9;9(6):e99391. doi: 10.1371/journal.pone.0099391. eCollection 2014. PLoS One. 2014. PMID: 24910998 Free PMC article.
References
- Narlikar G.J., Fan,H.Y. and Kingston,R.E. (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell, 108, 475–487. - PubMed
- Agalioti T., Lomvardas,S., Parekh,B., Yie,J., Maniatis,T. and Thanos,D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell, 103, 667–678. - PubMed
- Cosma M.P., Tanaka,T. and Nasmyth,K. (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell, 97, 299–311. - PubMed
- Cosma M. (2002) Ordered recruitment. Gene-specific mechanism of transcription activation. Mol. Cell, 10, 227. - PubMed
- Soutoglou E. and Talianidis,I. (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science, 295, 1901–1904. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous