The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts - PubMed (original) (raw)
The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts
Pulcherie Gueneau de Novoa et al. Nucleic Acids Res. 2004.
Abstract
tmRNA combines tRNA- and mRNA-like properties and ameliorates problems arising from stalled ribosomes. Research on the mechanism, structure and biology of tmRNA is served by the tmRNA website (http://www.indiana.edu/\~ tmrna), a collection of sequences, alignments, secondary structures and other information. Because many of these sequences are not in GenBank, a BLAST server has been added; another new feature is an abbreviated alignment for the tRNA-like domain only. Many tmRNA sequences from plastids have been added, five found in public sequence data and another 10 generated by direct sequencing; detection in early-branching members of the green plastid lineage brings coverage to all three primary plastid lineages. The new sequences include the shortest known tmRNA sequence. While bacterial tmRNAs usually have a lone pseudoknot upstream of the mRNA segment and a string of three or four pseudoknots downstream, plastid tmRNAs collectively show loss of pseudoknots at both postions. The pseudoknot-string region is also too short to contain the usual pseudoknot number in another new entry, the tmRNA sequence from a bacterial endosymbiont of insect cells, Tremblaya princeps. Pseudoknots may optimize tmRNA function in free-living bacteria, yet become dispensible when the endosymbiotic lifestyle relaxes selective pressure for fast growth.
Figures
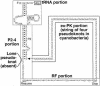
Figure 1
The shortest tmRNA, from the C.merolae plastid, showing the four portions analyzed in Table 1. The complete loss of the lone upstream pseudoknot is idiosyncratic.
Similar articles
- The tmRNA Website: invasion by an intron.
Williams KP. Williams KP. Nucleic Acids Res. 2002 Jan 1;30(1):179-82. doi: 10.1093/nar/30.1.179. Nucleic Acids Res. 2002. PMID: 11752287 Free PMC article. - Phylogenetic analysis of tmRNA secondary structure.
Williams KP, Bartel DP. Williams KP, et al. RNA. 1996 Dec;2(12):1306-10. RNA. 1996. PMID: 8972778 Free PMC article. - A third lineage with two-piece tmRNA.
Sharkady SM, Williams KP. Sharkady SM, et al. Nucleic Acids Res. 2004 Aug 23;32(15):4531-8. doi: 10.1093/nar/gkh795. Print 2004. Nucleic Acids Res. 2004. PMID: 15326226 Free PMC article. - Comparative sequence analysis of tmRNA.
Zwieb C, Wower I, Wower J. Zwieb C, et al. Nucleic Acids Res. 1999 May 15;27(10):2063-71. doi: 10.1093/nar/27.10.2063. Nucleic Acids Res. 1999. PMID: 10219077 Free PMC article. Review. - Role of horizontal gene transfer in the evolution of photosynthetic eukaryotes and their plastids.
Keeling PJ. Keeling PJ. Methods Mol Biol. 2009;532:501-15. doi: 10.1007/978-1-60327-853-9_29. Methods Mol Biol. 2009. PMID: 19271204 Review.
Cited by
- Comparative analysis of dinoflagellate chloroplast genomes reveals rRNA and tRNA genes.
Barbrook AC, Santucci N, Plenderleith LJ, Hiller RG, Howe CJ. Barbrook AC, et al. BMC Genomics. 2006 Nov 23;7:297. doi: 10.1186/1471-2164-7-297. BMC Genomics. 2006. PMID: 17123435 Free PMC article. - _Trans_-Translation Is an Appealing Target for the Development of New Antimicrobial Compounds.
Campos-Silva R, D'Urso G, Delalande O, Giudice E, Macedo AJ, Gillet R. Campos-Silva R, et al. Microorganisms. 2021 Dec 21;10(1):3. doi: 10.3390/microorganisms10010003. Microorganisms. 2021. PMID: 35056452 Free PMC article. Review. - Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB.
Hong SJ, Tran QA, Keiler KC. Hong SJ, et al. Mol Microbiol. 2005 Jul;57(2):565-75. doi: 10.1111/j.1365-2958.2005.04709.x. Mol Microbiol. 2005. PMID: 15978085 Free PMC article. - Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP.
Griffith KL, Grossman AD. Griffith KL, et al. Mol Microbiol. 2008 Nov;70(4):1012-25. doi: 10.1111/j.1365-2958.2008.06467.x. Epub 2008 Sep 22. Mol Microbiol. 2008. PMID: 18811726 Free PMC article. - Evolutionary patterns of non-coding RNAs.
Bompfünewerer AF, Flamm C, Fried C, Fritzsch G, Hofacker IL, Lehmann J, Missal K, Mosig A, Müller B, Prohaska SJ, Stadler BM, Stadler PF, Tanzer A, Washietl S, Witwer C. Bompfünewerer AF, et al. Theory Biosci. 2005 Apr;123(4):301-69. doi: 10.1016/j.thbio.2005.01.002. Theory Biosci. 2005. PMID: 18202870
References
- Keiler K.C., Waller,P.R. and Sauer,R.T. (1996) Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science, 271, 990–993. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials