Detection of homologous proteins by an intermediate sequence search - PubMed (original) (raw)
Comparative Study
Detection of homologous proteins by an intermediate sequence search
Bino John et al. Protein Sci. 2004 Jan.
Abstract
We developed a variant of the intermediate sequence search method (ISS(new)) for detection and alignment of weakly similar pairs of protein sequences. ISS(new) relates two query sequences by an intermediate sequence that is potentially homologous to both queries. The improvement was achieved by a more robust overlap score for a match between the queries through an intermediate. The approach was benchmarked on a data set of 2369 sequences of known structure with insignificant sequence similarity to each other (BLAST E-value larger than 0.001); 2050 of these sequences had a related structure in the set. ISS(new) performed significantly better than both PSI-BLAST and a previously described intermediate sequence search method. PSI-BLAST could not detect correct homologs for 1619 of the 2369 sequences. In contrast, ISS(new) assigned a correct homolog as the top hit for 121 of these 1619 sequences, while incorrectly assigning homologs for only nine targets; it did not assign homologs for the remainder of the sequences. By estimate, ISS(new) may be able to assign the folds of domains in approximately 29,000 of the approximately 500,000 sequences unassigned by PSI-BLAST, with 90% specificity (1 - false positives fraction). In addition, we show that the 15 alignments with the most significant BLAST E-values include the nearly best alignments constructed by ISS(new).
Figures
Figure 1.
An alignment of the target sequence with an intermediate and a putative homolog. The dotted and the dashed lines represent the intermediate aligned to the putative homolog and the target, respectively. The positions of the starting residues in the alignment of the intermediate with the putative homolog and the target are denoted by its and iqs, respectively. The positions of the ending residues are denoted by ite and iqe. The number of residues in the common overlap region of the intermediate is indicated by C.
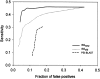
Figure 2.
Accuracy of ISSnew, ISSold, and PSI-BLAST on SEQS-EASY. The accuracy is described by the ROC curves (Materials and Methods) for PSI-BLAST (dashed line), ISSold (dotted line), and ISSnew (solid line).
Figure 3.
Accuracies of ISSnew, ISSold, and PSI-BLAST at different target-sequence lengths. (See Figure 2 ▶ legend for a description of the different symbols used.) The sequence lengths are less than 100 residues (A), between 100 and 200 residues (B), and greater than 200 residues (C).
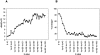
Figure 4.
Average alignment accuracy as a function of the thresholds on the overlap length (_x_-axis) and the ISSnew score (M). Error bars indicate the standard error of the mean; they are so small that they are almost invisible. Alignment accuracy is measured by the Cα RMSD between the compared structures (A) and coverage (B).
Figure 5.
Average alignment accuracy as a function of the _E_-value. The _E_-values were calculated using the BLOSUM62 amino acid substitution matrix. The alignments of the pairs in SEQS-HARD are obtained by ISSnew. (See Figure 4 ▶ for details.)
Figure 6.
Average alignment accuracy of the top five alignments selected by _E_-value as a function of the thresholds on the overlap length (_x_-axis) and the ISSnew score (M). The _E_-values were calculated using the BLOSUM62 amino acid substitution matrix. (See Figure 4 ▶ for details.)
Figure 7.
Average alignment accuracy of the best alignments in the selected set of alignments. Accuracy of the alignments selected by _E_-value using BC0030, BLOSUM62, and OPTIMA residue-type substitution matrices. Alignment accuracy is measured by the Cα RMSD of the structures (A) and coverage (B).
Similar articles
- Large-scale comparison of protein sequence alignment algorithms with structure alignments.
Sauder JM, Arthur JW, Dunbrack RL Jr. Sauder JM, et al. Proteins. 2000 Jul 1;40(1):6-22. doi: 10.1002/(sici)1097-0134(20000701)40:1<6::aid-prot30>3.0.co;2-7. Proteins. 2000. PMID: 10813826 - Efficient recognition of protein fold at low sequence identity by conservative application of Psi-BLAST: validation.
Stevens FJ. Stevens FJ. J Mol Recognit. 2005 Mar-Apr;18(2):139-49. doi: 10.1002/jmr.721. J Mol Recognit. 2005. PMID: 15558595 - Iterative sequence/secondary structure search for protein homologs: comparison with amino acid sequence alignments and application to fold recognition in genome databases.
Wallqvist A, Fukunishi Y, Murphy LR, Fadel A, Levy RM. Wallqvist A, et al. Bioinformatics. 2000 Nov;16(11):988-1002. doi: 10.1093/bioinformatics/16.11.988. Bioinformatics. 2000. PMID: 11159310 - Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements.
Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Schäffer AA, et al. Nucleic Acids Res. 2001 Jul 15;29(14):2994-3005. doi: 10.1093/nar/29.14.2994. Nucleic Acids Res. 2001. PMID: 11452024 Free PMC article. Review. - Getting the most from PSI-BLAST.
Jones DT, Swindells MB. Jones DT, et al. Trends Biochem Sci. 2002 Mar;27(3):161-4. doi: 10.1016/s0968-0004(01)02039-4. Trends Biochem Sci. 2002. PMID: 11893514 Review.
Cited by
- Detecting remotely related proteins by their interactions and sequence similarity.
Espadaler J, Aragüés R, Eswar N, Marti-Renom MA, Querol E, Avilés FX, Sali A, Oliva B. Espadaler J, et al. Proc Natl Acad Sci U S A. 2005 May 17;102(20):7151-6. doi: 10.1073/pnas.0500831102. Epub 2005 May 9. Proc Natl Acad Sci U S A. 2005. PMID: 15883372 Free PMC article. - ESG: extended similarity group method for automated protein function prediction.
Chitale M, Hawkins T, Park C, Kihara D. Chitale M, et al. Bioinformatics. 2009 Jul 15;25(14):1739-45. doi: 10.1093/bioinformatics/btp309. Epub 2009 May 12. Bioinformatics. 2009. PMID: 19435743 Free PMC article. - The limits of protein sequence comparison?
Pearson WR, Sierk ML. Pearson WR, et al. Curr Opin Struct Biol. 2005 Jun;15(3):254-60. doi: 10.1016/j.sbi.2005.05.005. Curr Opin Struct Biol. 2005. PMID: 15919194 Free PMC article. Review. - Graph pyramids for protein function prediction.
Sandhan T, Yoo Y, Choi J, Kim S. Sandhan T, et al. BMC Med Genomics. 2015;8 Suppl 2(Suppl 2):S12. doi: 10.1186/1755-8794-8-S2-S12. Epub 2015 May 29. BMC Med Genomics. 2015. PMID: 26044522 Free PMC article. - Profiles of Natural and Designed Protein-Like Sequences Effectively Bridge Protein Sequence Gaps: Implications in Distant Homology Detection.
Kumar G, Srinivasan N, Sandhya S. Kumar G, et al. Methods Mol Biol. 2022;2449:149-167. doi: 10.1007/978-1-0716-2095-3_5. Methods Mol Biol. 2022. PMID: 35507261
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
- Apostolico, A. and Giancarlo, R. 1998. Sequence alignment in molecular biology. J. Comput. Biol. 5 173–196. - PubMed
- Barton, G.J. 1994. Scop: Structural classification of proteins. Trends Biochem. Sci. 19 554–555. - PubMed
- Blake, J.D. and Cohen, F.E. 2001. Pairwise sequence alignment below the twilight zone. J. Mol. Biol. 307 721–735. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 GM062529/GM/NIGMS NIH HHS/United States
- R01 GM054762/GM/NIGMS NIH HHS/United States
- P50 GM62529/GM/NIGMS NIH HHS/United States
- R01 GM54762/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials