The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe - PubMed (original) (raw)
Comparative Study
. 2004 Jan 2;32(1):115-26.
doi: 10.1093/nar/gkh151. Print 2004.
Affiliations
- PMID: 14704348
- PMCID: PMC373264
- DOI: 10.1093/nar/gkh151
Comparative Study
The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe
Balazs Ribar et al. Nucleic Acids Res. 2004.
Abstract
The abasic (AP) sites, the major mutagenic and cytotoxic genomic lesions, induced directly by oxidative stress and indirectly after excision of damaged bases by DNA glycosylases, are repaired by AP-endonucleases (APEs). Among two APEs in Saccharomyces cerevisiae, Apn1 provides the major APE activity, and Apn2, the ortholog of the mammalian APE, provides back-up activity. We have cloned apn1 and apn2 genes of Schizosaccharomyces pombe, and have shown that inactivation of Apn2 and not Apn1 sensitizes this fission yeast to alkylation and oxidative damage-inducing agents, which is further enhanced by Apn1 inactivation. We also show that Uve1, present in S.pombe but not in S.cerevisiae, provides the back-up APE activity together with Apn1. We confirmed the presence of APE activity in recombinant Apn2 and in crude cell extracts. Thus S.pombe is distinct from S.cerevisiae, and is similar to mammalian cells in having Apn2 as the major APE.
Figures
Figure 1
Intron-containing region of the apn1 + (A) and apn2 + (B) genes. The intron sequences are shown in lower case. The consensus sequences for the 5′ splice, branch and 3′ splice sites are boxed [(17) and
http://www.sanger.ac.uk/Projects/S\_pombe
].
Figure 2
(A) Apn1 sequence alignment of S.pombe and S.cerevisiae Apn1with Nfo of E.coli. (B) Apn2 sequence alignment of S.pombe and S.cerevisiae, human APE2, human APE1 and E.coli Xth proteins. Identical and highly conserved residues are indicated in red. Spaces indicate gaps introduced to optimize alignment by the MultiAlin program (23):
http://prodes.toulouse.inra.fr/multalin/multalin.html
. GenBank accession numbers AY483157 for apn1+ and AY483158 for apn2+.
Figure 2
(A) Apn1 sequence alignment of S.pombe and S.cerevisiae Apn1with Nfo of E.coli. (B) Apn2 sequence alignment of S.pombe and S.cerevisiae, human APE2, human APE1 and E.coli Xth proteins. Identical and highly conserved residues are indicated in red. Spaces indicate gaps introduced to optimize alignment by the MultiAlin program (23):
http://prodes.toulouse.inra.fr/multalin/multalin.html
. GenBank accession numbers AY483157 for apn1+ and AY483158 for apn2+.
Figure 3
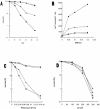
(A) MMS sensitivity of _apn1_Δ and _apn2_Δ strains. Cells grown overnight in YEL medium were treated with 0.2% MMS for various times. (B) MMS-induced mutagenesis at the can1 gene of S.pombe. The details are described in Materials and Methods. (C) Phleomycin D1 toxicity. Cells grown overnight in YEL medium were treated with phleomycin D1 for 1 h before spreading on YEA plates. (D) UV sensitivity of _S.pombe apn1_Δ and _apn2_Δ mutant strains. Sensitivity of the strains to UV was measured after exposure to various UV doses. PO29, wild type (diamonds); PO61, _apn1_Δ (inverted triangles); PO24, _apn2_Δ (upright triangles); PO60, _apn1_Δ_apn2_Δ (circles).
Figure 4
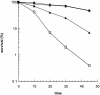
Effect of _uve1_Δ mutation on MMS sensitivity of the _apn2_Δ mutant. Other details are as in Figure 3. PO29, wild type (diamonds); PO24, _apn2_Δ (triangles); PO89, _uve1_Δ (open circles); PO90, _apn2_Δ_uve1_Δ (open squares).
Figure 5
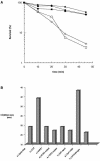
(A) Complementation of MMS sensitivity of the _apn1_Δ_apn2_Δ (PO60) strain with wild-type S.pombe Apn2-His6 (pNBR110) (filled squares); human APE1 (pRBR113) (filled circles); and human APE2 (pNBR103) (open squares); wild-type control, leu1-32 (PO4), with control CAT expression vector (pNMT1-CAT; Invitrogen) (filled diamonds); sensitivity of _apn1_Δ_apn2_Δ: (PO60) transformed with pNMT1-CAT (open diamonds). Other details are described in Materials and Methods. (B) Agar diffusion assay of complementation of _apn1_Δ_apn2_Δ (PO60) and wild-type (PO4) strains as described in Materials and Methods. Data represent results from an average of three or more experiments. (a) Wild-type Apn2-His6 on plasmid pNBR110 in PO60; (b) control pNMT1-CAT plasmid in PO60; (c) human APE1 on plasmid pRBR113 in PO60; (d) Apn2-A402A403 mutation (PCNA) on plasmid pNBR114 in PO60; (e) Apn2-A456A457A458 mutation on plasmid pNBR115 in PO60; (f) wild-type GST–Apn2 on plasmid pEBR116 in PO60; (g) wild-type Apn1-His6 on plasmid pNBR117 in PO60; (h) pNMT1-CAT plasmid in PO4
Figure 6
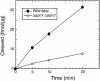
(A) Purification of GST–Apn2. Eluted GST–Apn2 fusion protein was analyzed on a 7.5% denaturing polyacrylamide gel and stained with Coomassie blue. Lane 1, molecular weight standards; lane 2, 3 µg of eluted GST–Apn2 protein. (B) Kinetics of APE activity of GST–Apn2; lane (–) is the no-protein control. The details are described in Materials and Methods.
Figure 7
Analysis of APE activity in S.pombe extract. Wild-type and mutant cells were broken by French press and centrifuged. Crude cell extract (1 µg) was mixed with 50 fmol of the oligonucleotide containing the AP site in reaction mixture (20 µl) containing 20 mM Tris pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT and 100 µg/ml BSA. After incubation at 37°C, the 5′-labeled 30mer product was identified with PhosphorImager (Molecular Dynamics) after electrophoretic separation on 20% polyacrylamide gels containing 7 M urea (26,45).
Similar articles
- A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe.
Alseth I, Korvald H, Osman F, Seeberg E, Bjørås M. Alseth I, et al. Nucleic Acids Res. 2004 Sep 27;32(17):5119-25. doi: 10.1093/nar/gkh851. Print 2004. Nucleic Acids Res. 2004. PMID: 15452279 Free PMC article. - Schizosaccharomyces pombe encodes a mutated AP endonuclease 1.
Laerdahl JK, Korvald H, Nilsen L, Dahl-Michelsen K, Rognes T, Bjørås M, Alseth I. Laerdahl JK, et al. DNA Repair (Amst). 2011 Mar 7;10(3):296-305. doi: 10.1016/j.dnarep.2010.11.014. Epub 2010 Dec 28. DNA Repair (Amst). 2011. PMID: 21193357 - Use of yeast for detection of endogenous abasic lesions, their source, and their repair.
Boiteux S, Guillet M. Boiteux S, et al. Methods Enzymol. 2006;408:79-91. doi: 10.1016/S0076-6879(06)08006-2. Methods Enzymol. 2006. PMID: 16793364 - Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae.
Boiteux S, Guillet M. Boiteux S, et al. DNA Repair (Amst). 2004 Jan 5;3(1):1-12. doi: 10.1016/j.dnarep.2003.10.002. DNA Repair (Amst). 2004. PMID: 14697754 Review. - Regulation of eukaryotic abasic endonucleases and their role in genetic stability.
Demple B, Harrison L, Wilson DM 3rd, Bennett RA, Takagi T, Ascione AG. Demple B, et al. Environ Health Perspect. 1997 Jun;105 Suppl 4(Suppl 4):931-4. doi: 10.1289/ehp.97105s4931. Environ Health Perspect. 1997. PMID: 9255583 Free PMC article. Review.
Cited by
- Base Excision Repair AP-Endonucleases-Like Genes Modulate DNA Damage Response and Virulence of the Human Pathogen Cryptococcus neoformans.
Oliveira RKM, Hurtado FA, Gomes PH, Puglia LL, Ferreira FF, Ranjan K, Albuquerque P, Poças-Fonseca MJ, Silva-Pereira I, Fernandes L. Oliveira RKM, et al. J Fungi (Basel). 2021 Feb 12;7(2):133. doi: 10.3390/jof7020133. J Fungi (Basel). 2021. PMID: 33673204 Free PMC article. - A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe.
Alseth I, Korvald H, Osman F, Seeberg E, Bjørås M. Alseth I, et al. Nucleic Acids Res. 2004 Sep 27;32(17):5119-25. doi: 10.1093/nar/gkh851. Print 2004. Nucleic Acids Res. 2004. PMID: 15452279 Free PMC article. - Early Steps in the DNA Base Excision Repair Pathway of a Fission Yeast Schizosaccharomyces pombe.
Kanamitsu K, Ikeda S. Kanamitsu K, et al. J Nucleic Acids. 2010 Sep 16;2010:450926. doi: 10.4061/2010/450926. J Nucleic Acids. 2010. PMID: 20936170 Free PMC article. - Two essential but distinct functions of the mammalian abasic endonuclease.
Izumi T, Brown DB, Naidu CV, Bhakat KK, Macinnes MA, Saito H, Chen DJ, Mitra S. Izumi T, et al. Proc Natl Acad Sci U S A. 2005 Apr 19;102(16):5739-43. doi: 10.1073/pnas.0500986102. Epub 2005 Apr 11. Proc Natl Acad Sci U S A. 2005. PMID: 15824325 Free PMC article. - Mitochondrial DNA repair and association with aging--an update.
Gredilla R, Bohr VA, Stevnsner T. Gredilla R, et al. Exp Gerontol. 2010 Aug;45(7-8):478-88. doi: 10.1016/j.exger.2010.01.017. Epub 2010 Jan 22. Exp Gerontol. 2010. PMID: 20096766 Free PMC article. Review.
References
- Breen A.P. and Murphy,J.A. (1995) Reactions of oxyl radicals with DNA. Free Radical Biol. Med., 18, 1033–1077. - PubMed
- Mitra S., Hazra,T.K., Roy,R., Ikeda,S., Biswas,T., Lock,J., Boldogh,I. and Izumi,T. (1997) Complexities of DNA base excision repair in mammalian cells. Mol. Cell, 7, 305–312. - PubMed
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. - PubMed
- Nakamura J. and Swenberg,J.A. (1999) Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res., 59, 2522–2536. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P50 ES06676/ES/NIEHS NIH HHS/United States
- ES08457/ES/NIEHS NIH HHS/United States
- R01 CA53791/CA/NCI NIH HHS/United States
- R01 ES008457/ES/NIEHS NIH HHS/United States
- R01 CA053791/CA/NCI NIH HHS/United States
- P30 ES006676/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous