A model of the ACE2 structure and function as a SARS-CoV receptor - PubMed (original) (raw)
A model of the ACE2 structure and function as a SARS-CoV receptor
Ponraj Prabakaran et al. Biochem Biophys Res Commun. 2004.
Abstract
The angiotensin-converting enzyme 2 (ACE2) is an important regulator of the renin-angiotensin system and was very recently identified as a functional receptor for the SARS virus. The ACE2 sequence is similar (sequence identities 43% and 35%, and similarities 61% and 55%, respectively) to those of the testis-specific form of ACE (tACE) and the Drosophila homolog of ACE (AnCE). The high level of sequence similarity allowed us to build a robust homology model of the ACE2 structure with a root-mean-square deviation from the aligned crystal structures of tACE and AnCE less than 0.5A. A prominent feature of the model is a deep channel on the top of the molecule that contains the catalytic site. Negatively charged ridges surrounding the channel may provide a possible binding site for the positively charged receptor-binding domain (RBD) of the S-glycoprotein, which we recently identified [Biochem. Biophys. Res. Commun. 312 (2003) 1159]. Several distinct patches of hydrophobic residues at the ACE2 surface were noted at close proximity to the charged ridges that could contribute to binding. These results suggest a possible binding region for the SARS-CoV S-glycoprotein on ACE2 and could help in the design of experiments to further elucidate the structure and function of ACE2.
Figures
Fig. 1
Multiple sequence alignment of ACE2, tACE, and AnCE using CLUSTALW. The sequence numbering is the same as in the crystal structures. The N-glycosylation sites are underlined; putative binding residues in ACE2 are in boldface letters and boxed along with the corresponding aligned residues in tACE and AnCE. The identical and similar residues are shown in black and gray backgrounds, respectively.
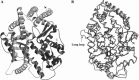
Fig. 2
The model of the ACE2 structure. (A) A ribbon representation of the ACE2 model. The N- and C-termini are indicated. (B) Superposition of the ACE2 model structure with the crystal structures of tACE and AnCE based on the Cα-atoms of ACE2, tACE, and AnCE (ACE2, dark gray; tACE, light gray; and AnCE, black). The long loop inserted between N210 and Q221 that is unique for ACE2 is indicated.
Fig. 3
Analysis of the ACE2 model structure. (A) Representation of negative and positive molecular surface potentials in red and blue, respectively. The channel at the top of the molecule containing the catalytic site and the surrounding ridges containing negatively charged residues is indicated. (B) Distribution of glycosylation sites (green) on the ACE2 surface. (C) The backbone warm representing the orientation of the main chain. (D) Distribution of hydrophobic patches on the ACE2 surface.
Fig. 4
Solvent accessible surface areas (right column, in Å2) for ACE2 amino acid residues that are significantly exposed to solvent at the surface of the molecule. The cut-off for significant surface exposure here is assumed to be 45% ratio value defined as the ratio of side-chain surface area to a “random coil” value per residue in the tripeptide Gly–X–Gly. The middle column represents the amino acid residue number.
Fig. 5
Conservation of amino acid residues in human ACE2, human ACE, and mouse ACE2 that could contribute to interactions with the _S_-glycoprotein. Identities are marked by a pipe (|), highly conservative replacements by a colon (:), and replacements with lower scores by a dot (·). The numbers denote the amino acid residue positions in the sequence. Note that the similarity of these ACE2 residues with the corresponding residues of mouse ACE2 is much higher than with the respective human ACE residues.
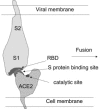
Fig. 6
Schematic representation of the interaction between ACE2 and the SARS-CoV _S_-glycoprotein leading to binding and fusion. The RBD is depicted as a surface containing a cavity(s) that binds a ridge(s) close to the deep channel containing the catalytic site.
Similar articles
- A molecular docking model of SARS-CoV S1 protein in complex with its receptor, human ACE2.
Zhang Y, Zheng N, Hao P, Cao Y, Zhong Y. Zhang Y, et al. Comput Biol Chem. 2005 Jun;29(3):254-7. doi: 10.1016/j.compbiolchem.2005.04.008. Comput Biol Chem. 2005. PMID: 15979045 Free PMC article. - Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Lan J, et al. Nature. 2020 May;581(7807):215-220. doi: 10.1038/s41586-020-2180-5. Epub 2020 Mar 30. Nature. 2020. PMID: 32225176 - Structure of SARS coronavirus spike receptor-binding domain complexed with receptor.
Li F, Li W, Farzan M, Harrison SC. Li F, et al. Science. 2005 Sep 16;309(5742):1864-8. doi: 10.1126/science.1116480. Science. 2005. PMID: 16166518 - Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus.
Kuhn JH, Li W, Choe H, Farzan M. Kuhn JH, et al. Cell Mol Life Sci. 2004 Nov;61(21):2738-43. doi: 10.1007/s00018-004-4242-5. Cell Mol Life Sci. 2004. PMID: 15549175 Free PMC article. Review. - Membrane-associated zinc peptidase families: comparing ACE and ACE2.
Guy JL, Lambert DW, Warner FJ, Hooper NM, Turner AJ. Guy JL, et al. Biochim Biophys Acta. 2005 Aug 1;1751(1):2-8. doi: 10.1016/j.bbapap.2004.10.010. Epub 2004 Nov 6. Biochim Biophys Acta. 2005. PMID: 16054014 Free PMC article. Review.
Cited by
- Silver and Carbon Nanomaterials/Nanocomplexes as Safe and Effective ACE2-S Binding Blockers on Human Skin Cell Lines.
Hotowy A, Strojny-Cieślak B, Ostrowska A, Zielińska-Górska M, Kutwin M, Wierzbicki M, Sosnowska M, Jaworski S, Chwalibóg A, Kotela I, Sawosz Chwalibóg E. Hotowy A, et al. Molecules. 2024 Jul 29;29(15):3581. doi: 10.3390/molecules29153581. Molecules. 2024. PMID: 39124987 Free PMC article. - DNA-based assay for calorimetric determination of protein concentrations in pure or mixed solutions.
Eskew MW, Reardon P, Benight AS. Eskew MW, et al. PLoS One. 2024 Mar 1;19(3):e0298969. doi: 10.1371/journal.pone.0298969. eCollection 2024. PLoS One. 2024. PMID: 38427623 Free PMC article. - Binding behavior of receptor binding domain of the SARS-CoV-2 virus and ivermectin.
Gossen KR, Zhang M, Nikolov ZL, Fernando SD, King MD. Gossen KR, et al. Sci Rep. 2024 Feb 2;14(1):2743. doi: 10.1038/s41598-024-53086-0. Sci Rep. 2024. PMID: 38302638 Free PMC article. - ACE2 knockout hinders SARS-CoV-2 propagation in iPS cell-derived airway and alveolar epithelial cells.
Niwa R, Sakai K, Lung MSY, Matsumoto T, Mikawa R, Maehana S, Suzuki M, Yamamoto Y, Maurissen TL, Hirabayashi A, Noda T, Kubo M, Gotoh S, Woltjen K. Niwa R, et al. Front Cell Dev Biol. 2023 Dec 8;11:1290876. doi: 10.3389/fcell.2023.1290876. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 38149046 Free PMC article. - Synthetic graphene-copper nanocomposites interact with the hACE-2 enzyme and inhibit its biochemical activity.
Sulthana S, Bhatti A, Mathew E, Quazi SH, Gaudreault NN, DeLong R, Aryal S. Sulthana S, et al. Nanoscale Adv. 2023 Nov 10;6(1):188-196. doi: 10.1039/d3na00468f. eCollection 2023 Dec 19. Nanoscale Adv. 2023. PMID: 38125590 Free PMC article.
References
- Dimitrov D.S. Cell biology of virus entry. Cell. 2000;101:697–702. - PubMed
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., Breitbart R.E., Acton S. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000;87:E1–E9. - PubMed
- Tipnis S.R., Hooper N.M., Hyde R., Karran E., Christie G., Turner A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000;275:33238–33243. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous