Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes - PubMed (original) (raw)
Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes
Keigo Machida et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Hepatitis C virus (HCV) is a nonretroviral oncogenic RNA virus, which is frequently associated with hepatocellular carcinoma (HCC) and B cell lymphoma. We demonstrated here that acute and chronic HCV infection caused a 5- to 10-fold increase in mutation frequency in Ig heavy chain, BCL-6, p53, and beta-catenin genes of in vitro HCV-infected B cell lines and HCV-associated peripheral blood mononuclear cells, lymphomas, and HCCs. The nucleotide-substitution pattern of p53 and beta-catenin was different from that of Ig heavy chain in HCV-infected cells, suggesting two different mechanisms of mutation. In addition, the mutated protooncogenes were amplified in HCV-associated lymphomas and HCCs, but not in lymphomas of nonviral origin or HBV-associated HCC. HCV induced error-prone DNA polymerase zeta, polymerase iota, and activation-induced cytidine deaminase, which together, contributed to the enhancement of mutation frequency, as demonstrated by the RNA interference experiments. These results indicate that HCV induces a mutator phenotype and may transform cells by a hit-and-run mechanism. This finding provides a mechanism of oncogenesis for an RNA virus.
Figures
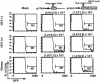
Fig. 1.
Determination of mutation frequencies by using reporter GFP genes in B cells. Cloned Raji cells infected with HCV or UV-irradiated HCV [HCV(-)] were transfected with the indicated reporter plasmid (pI-TAA or pI-Enh) on day 8 after infection (11). pI-Enh reporter plasmids without the enhancer element were used as the control. The cells were analyzed for expression of GFP by fluorescence-activated cell sorting analysis. Frequencies of mutations are shown.
Fig. 2.
LM-PCR amplification of DSBs in HCV-infected and control Raji cells. (A_–_C) Blunt-ended DSBs were detected over the entire genome (A) or specifically in the Ig (B) or p53 (C). Raji cells were infected with HCV or UV-irradiated HCV [HCV(-)] and were examined at different days after infection. HCV(-) cells were infected with UV-irradiated HCV and are negative for HCV RNA. MK, DNA size marker; H2O, water control. (D) PCR amplification of the VH region was used as a loading control. (E) (M) HCV-RNA was detected by RT-PCR. (F) DSBs in HCV-infected JT cells were compared with those in the infected Raji cells. (G) Cell growth curve of HCV-infected Raji cells as determined by Trypan blue exclusion assay. (H) Semiquantitative RT-PCR detection of error-prone DNA polymerases and AID in Raji cells. RNA transcripts of various polymerases and the AID gene were reverse-transcribed in Raji cells on day 8 after HCV infection. cDNA of different dilutions (no dilution, 1:5, and 1:25) were then used for PCR amplification. Ramos cells were included as a control. (I) RNA samples from Raji cells on different days after HCV infection were used for amplification of polymerase ζ or polymerase ι, and β-actin in the same reaction. (J) Northern blots for AID and GAPDH in Raji cells on different days after infection. The relative amounts of the AID transcript on different days after infection were quantified by an image analyzer and are indicated below. (K) RT-PCR of AID in HCV-infected and non-infected PBMC from 3–4 individual HCV patients and healthy individuals. Three different concentrations of cDNA from each PBMC sample were used. RT-PCR of β-actin served as internal control. MK, DNA size marker, H2O, water control. (L) Fold induction of AID was normalized with the expression of β-actin.
Fig. 3.
(A and B) The effect of siRNA on the levels of AID and DNA polymerase ι during HCV infection. Western blot analysis shows AID and error-prone polymerases expression at 8-days after transfection in HCV-infected cells in the presence of the specific or scramble siRNA. Western blot of β-actin was used as loading control. AID-/- or polymeraseι-/- BL2 cells served as negative control. HCV-RNA was detected with RT-PCR. (C) Antisense oligodeoxynucleotides against polymerase ζ were used as reported (23). SCR, scramble oligonucleotide; AS, antisense oligonucleotide against polymerase ζ.(D) HCV RNA was detected with RT-PCR.
Similar articles
- Tumor-suppressor p53 gene in hepatitis C and B virus-associated human hepatocellular carcinoma.
Shieh YS, Nguyen C, Vocal MV, Chu HW. Shieh YS, et al. Int J Cancer. 1993 Jun 19;54(4):558-62. doi: 10.1002/ijc.2910540407. Int J Cancer. 1993. PMID: 8390407 - Hepatitis C virus infection activates the immunologic (type II) isoform of nitric oxide synthase and thereby enhances DNA damage and mutations of cellular genes.
Machida K, Cheng KT, Sung VM, Lee KJ, Levine AM, Lai MM. Machida K, et al. J Virol. 2004 Aug;78(16):8835-43. doi: 10.1128/JVI.78.16.8835-8843.2004. J Virol. 2004. PMID: 15280491 Free PMC article. - Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis.
Hsu HC, Jeng YM, Mao TL, Chu JS, Lai PL, Peng SY. Hsu HC, et al. Am J Pathol. 2000 Sep;157(3):763-70. doi: 10.1016/s0002-9440(10)64590-7. Am J Pathol. 2000. PMID: 10980116 Free PMC article. - Hepatocarcinogenesis: hepatitis viruses and altered tumor suppressor gene function.
Tabor E. Tabor E. Princess Takamatsu Symp. 1995;25:151-61. Princess Takamatsu Symp. 1995. PMID: 8875620 Review. - Heterogeneity and coexistence of oncogenic mechanisms involved in HCV-associated B-cell lymphomas.
Carloni G, Fioretti D, Rinaldi M, Ponzetto A. Carloni G, et al. Crit Rev Oncol Hematol. 2019 Jun;138:156-171. doi: 10.1016/j.critrevonc.2019.04.005. Epub 2019 Apr 16. Crit Rev Oncol Hematol. 2019. PMID: 31092372 Review.
Cited by
- Relationship between codon 249 mutation in exon 7 of p53 gene and diagnosis of hepatocellular carcinoma.
El-Din HG, Ghafar NA, Saad NE, Aziz M, Rasheed D, Hassan EM. El-Din HG, et al. Arch Med Sci. 2010 Jun 30;6(3):348-55. doi: 10.5114/aoms.2010.14254. Arch Med Sci. 2010. PMID: 22371770 Free PMC article. - Repair of DNA strand breaks in a minichromosome in vivo: kinetics, modeling, and effects of inhibitors.
Kumala S, Fujarewicz K, Jayaraju D, Rzeszowska-Wolny J, Hancock R. Kumala S, et al. PLoS One. 2013;8(1):e52966. doi: 10.1371/journal.pone.0052966. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23382828 Free PMC article. - Rev1, Rev3, or Rev7 siRNA Abolishes Ultraviolet Light-Induced Translesion Replication in HeLa Cells: A Comprehensive Study Using Alkaline Sucrose Density Gradient Sedimentation.
Takezawa J, Ishimi Y, Aiba N, Yamada K. Takezawa J, et al. J Nucleic Acids. 2010 Dec 1;2010:750296. doi: 10.4061/2010/750296. J Nucleic Acids. 2010. PMID: 21151666 Free PMC article. - Direct effects of hepatitis C virus on the lymphoid cells.
Kondo Y, Shimosegawa T. Kondo Y, et al. World J Gastroenterol. 2013 Nov 28;19(44):7889-95. doi: 10.3748/wjg.v19.i44.7889. World J Gastroenterol. 2013. PMID: 24307783 Free PMC article. Review. - Hepatitis C virus infection of T cells inhibits proliferation and enhances fas-mediated apoptosis by down-regulating the expression of CD44 splicing variant 6.
Kondo Y, Machida K, Liu HM, Ueno Y, Kobayashi K, Wakita T, Shimosegawa T, Lai MM. Kondo Y, et al. J Infect Dis. 2009 Mar 1;199(5):726-36. doi: 10.1086/596739. J Infect Dis. 2009. PMID: 19199548 Free PMC article.
References
- Ferri, C., Caracciolo, F., Zignego, A. L., La Civita, L., Monti, M., Longombardo, G., Lombardini, F., Greco, F., Capochiani, E., Mazzoni, A., et al. (1994) Br. J. Haematol. 88, 392-394. - PubMed
- Teramoto, T., Satonaka, K., Kitazawa, S., Fujimori, T., Hayashi, K. & Maeda, S. (1994) Cancer Res. 54, 231-235. - PubMed
- Takeda, S., Shibata, M., Morishima, T., Harada, A., Nakao, A., Takagi, H. & Nagai, Y. (1992) Cancer 70, 2255-2259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous