RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly - PubMed (original) (raw)
RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly
Sowmya Swaminathan et al. J Cell Biol. 2004.
Abstract
The RanGTPase activating protein RanGAP1 has essential functions in both nucleocytoplasmic transport and mitosis. In interphase, a significant fraction of vertebrate SUMO1-modified RanGAP1 forms a stable complex with the nucleoporin RanBP2/Nup358 at nuclear pore complexes. RanBP2 not only acts in the RanGTPase cycle but also is a SUMO1 E3 ligase. Here, we show that RanGAP1 is phosphorylated on residues T409, S428, and S442. Phosphorylation occurs before nuclear envelope breakdown and is maintained throughout mitosis. Nocodazole arrest leads to quantitative phosphorylation. The M-phase kinase cyclin B/Cdk1 phosphorylates RanGAP1 efficiently in vitro, and T409 phosphorylation correlates with nuclear accumulation of cyclin B1 in vivo. We find that phosphorylated RanGAP1 remains associated with RanBP2/Nup358 and the SUMO E2-conjugating enzyme Ubc9 in mitosis, hence mitotic phosphorylation may have functional consequences for the RanGTPase cycle and/or for RanBP2-dependent sumoylation.
Figures
Figure 1.
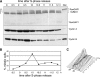
Cell cycle–dependent phosphorylation of RanGAP1. (A) HeLa cells arrested in S-phase with thymidine were released from the S-phase block, harvested at the indicated times, and analyzed by immunoblotting with antibodies to RanGAP1, cyclin A, and cyclin B. For optimal resolution of RanGAP1 species a, b, and c, 6% SDS-PAGE was used for the top panel. Note that all time points were run on the same gel. (B) Mitotic index was determined by visually scoring fixed cells stained with Hoechst for the presence of condensed chromosomes. About 200 cells were scored at each time point. (C) Cell cycle analysis: DNA content of each sample was determined by propidium iodine staining and analysis by flow cytometry.
Figure 2.
Mitotic RanGAP1 is phosphorylated at residues T409, S428, and S442 in vivo. (A) RanGAP1 species required for mass spectrometry (indicated as bands a, b, and c) were enriched by immunoprecipitation from HeLa cell extracts. A fraction of the precipitates was subjected to immunoblotting with α-RanGAP1 antibodies. The remainder was separated on SDS-PAGE (not depicted), and Coomassie-stained bands a, b, and c were excised from lanes 2, 3, and 1, respectively. cyc, cycling cells; nocodazole, nocodazole-arrested cells; dig, digitonin lysis; hyp, hypotonic swelling extracts. (B) MALDI-TOF mass spectra of a tryptic digest of RanGAP1 species enriched as described in A. Top, band c; middle, band a; bottom, band b. A peptide (amino acids 407–445 in human RanGAP1) containing up to three phosphates was identified. A selected mass range containing different phosphorylation stages (0, 1, 2, and 3 Ph) of this peptide is depicted. (C) Scheme showing the localization of two phosphorylation sites through fragmentation of the doubly phosphorylated peptide (amino acids 414–445) of RanGAP1 using an electrospray ionization ion trap mass spectrometer. The number of phosphorylation sites that were observed on each fragment is added as an arabic number to the arrows. (D) RanGAP1 immunoprecipitated with α RanGAP1 antibodies from cycling and nocodazole-arrested HeLa cells was analyzed by immunoblotting with the indicated phosphospecific antibodies. (E) All three phosphorylation sites of human RanGAP1 are conserved in mouse, areas surrounding S442 and S428 are also conserved in Xenopus laevis RanGAP1.
Figure 3.
RanGAP1 phosphorylation takes place before nuclear envelope breakdown. (A) HeLa cells fixed and permeabilized with 4% PFA and 0.2% Triton X-100 were subjected to indirect immunofluorescence with α p-T409 antibody. (B) HeLa cells were fixed with 2% PFA, permeabilized with 0.2% Triton X-100, and stained with primary antibodies as indicated. (C) At least three distinct phosphorylated RanGAP1 species exist during mitosis. (lanes 2–7) α RanGAP1 immunoprecipitates from cells harvested at indicated times after release from thymidine (same cells as in Fig. 1). (lanes 1, 8, and 9) Total extracts from cycling (cyc) and nocodazole (noc) extracts served as controls. Immunoblotting was done with the indicated phosphospecific antibodies.
Figure 4.
RanGAP1 phosphorylation by Cdks. (A) Phosphorylation of RanGAP1 in HeLa cell extracts is inhibited by p27. Recombinant sumoylated RanGAP1 was incubated with nocodazole extracts in the presence of ATP and analyzed by immunoblotting with α RanGAP1. Phosphorylation results in a mobility shift (arrows). Where indicated, extracts were preincubated with p27 before RanGAP1*SUMO1 addition. (B) In vitro phosphorylation of recombinant RanGAP1 with cyclin A/Cdk2 or cyclin B/Cdk1. After incubation, samples were analyzed by SDS-PAGE and autoradiography. p27 was included to demonstrate specificity of the kinase preparations. Mouse RanGAP1 mutants: T-D, RanGAP1-T411D; S-D, RanGAP1-S444D; DD, RanGAP1-T411D, S444D. (C) In vivo phosphorylation of human RanGAP1 T409 correlates with levels and localization of cyclin B. HeLa cells fixed with 2% PFA and permeabilized with 0.2% Triton X-100 were stained with α p-T409 antibodies and α cyclin B.
Figure 5.
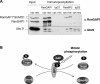
RanGAP1*SUMO1 phosphoforms a and b remain associated with RanBP2 and Ubc9 in nocodazole-arrested HeLa cells. (A) α RanGAP1 and α RanBP2 immunoprecipitates from cycling and nocodazole-arrested cells (RIPA extracts) were analyzed for the presence of RanGAP1 (top) and Ubc9 (bottom) by immunoblotting. IgGs 1 and 2 were from the respective preimmune sera and served as specificity controls. (B) Model: the RanGAP1*SUMO1–RanBP2 complex serves a dual function as an activator of Ran-GTP hydrolysis and as a SUMO E3 ligase. Either process may be affected by mitotic phosphorylation of RanGAP1.
Similar articles
- The RanBP2/RanGAP1*SUMO1/Ubc9 complex is a multisubunit SUMO E3 ligase.
Werner A, Flotho A, Melchior F. Werner A, et al. Mol Cell. 2012 May 11;46(3):287-98. doi: 10.1016/j.molcel.2012.02.017. Epub 2012 Mar 29. Mol Cell. 2012. PMID: 22464730 - The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes.
Ritterhoff T, Das H, Hofhaus G, Schröder RR, Flotho A, Melchior F. Ritterhoff T, et al. Nat Commun. 2016 May 10;7:11482. doi: 10.1038/ncomms11482. Nat Commun. 2016. PMID: 27160050 Free PMC article. - Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2.
Gareau JR, Reverter D, Lima CD. Gareau JR, et al. J Biol Chem. 2012 Feb 10;287(7):4740-51. doi: 10.1074/jbc.M111.321141. Epub 2011 Dec 22. J Biol Chem. 2012. PMID: 22194619 Free PMC article. - Sumoylating and desumoylating enzymes at nuclear pores: underpinning their unexpected duties?
Palancade B, Doye V. Palancade B, et al. Trends Cell Biol. 2008 Apr;18(4):174-83. doi: 10.1016/j.tcb.2008.02.001. Epub 2008 Mar 3. Trends Cell Biol. 2008. PMID: 18313922 Review. - Adding pieces to the puzzling plant nuclear envelope.
Meier I, Brkljacic J. Meier I, et al. Curr Opin Plant Biol. 2009 Dec;12(6):752-9. doi: 10.1016/j.pbi.2009.09.016. Epub 2009 Oct 28. Curr Opin Plant Biol. 2009. PMID: 19875325 Review.
Cited by
- Regulation of T cell proliferation by JMJD6 and PDGF-BB during chronic hepatitis B infection.
Chen CF, Feng X, Liao HY, Jin WJ, Zhang J, Wang Y, Gong LL, Liu JJ, Yuan XH, Zhao BB, Zhang D, Chen GF, Wan Y, Guo J, Yan HP, He YW. Chen CF, et al. Sci Rep. 2014 Sep 15;4:6359. doi: 10.1038/srep06359. Sci Rep. 2014. PMID: 25219359 Free PMC article. - SCFFbxw5 mediates transient degradation of actin remodeller Eps8 to allow proper mitotic progression.
Werner A, Disanza A, Reifenberger N, Habeck G, Becker J, Calabrese M, Urlaub H, Lorenz H, Schulman B, Scita G, Melchior F. Werner A, et al. Nat Cell Biol. 2013 Feb;15(2):179-88. doi: 10.1038/ncb2661. Epub 2013 Jan 13. Nat Cell Biol. 2013. PMID: 23314863 Free PMC article. - Visualization of human karyopherin beta-1/importin beta-1 interactions with protein partners in mitotic cells by co-immunoprecipitation and proximity ligation assays.
Di Francesco L, Verrico A, Asteriti IA, Rovella P, Cirigliano P, Guarguaglini G, Schininà ME, Lavia P. Di Francesco L, et al. Sci Rep. 2018 Jan 30;8(1):1850. doi: 10.1038/s41598-018-19351-9. Sci Rep. 2018. PMID: 29382863 Free PMC article. - The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent nuclear import.
Hutten S, Flotho A, Melchior F, Kehlenbach RH. Hutten S, et al. Mol Biol Cell. 2008 May;19(5):2300-10. doi: 10.1091/mbc.e07-12-1279. Epub 2008 Feb 27. Mol Biol Cell. 2008. PMID: 18305100 Free PMC article. - RanBP2/Nup358 Mediates Sumoylation of STAT1 and Antagonizes Interferon-α-Mediated Antiviral Innate Immunity.
Li J, Su L, Jiang J, Wang YE, Ling Y, Qiu Y, Yu H, Huang Y, Wu J, Jiang S, Zhang T, Palazzo AF, Shen Q. Li J, et al. Int J Mol Sci. 2023 Dec 25;25(1):299. doi: 10.3390/ijms25010299. Int J Mol Sci. 2023. PMID: 38203469 Free PMC article.
References
- Arnaoutov, A., and M. Dasso. 2003. The Ran GTPase regulates kinetochore function. Dev. Cell. 5:99–111. - PubMed
- Bonifacino, J.S., M. Dasso, J. Lippincott-Schwartz, J.B. Harford, and K.M. Yamada. 1999. Current Protocols in Cell Biology. John Wiley and Sons, New York. Also available at http://www.mrw2.interscience.wiley.com/cponline.
- Favreau, C., H.J. Worman, R.W. Wozniak, T. Frappier, and J.C. Courvalin. 1996. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein Gp210. Biochemistry. 35:8035–8044. - PubMed
- Gobom, J., E. Nordhoff, E. Mirgorodskaya, R. Ekman, and P. Roepstorff. 1999. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34:105–116. - PubMed
- Guarguaglini, G., L. Renzi, F. D'Ottavio, B. Di Fiore, M. Casenghi, E. Cundari, and P. Lavia. 2000. Regulated Ran-binding protein 1 activity is required for organization and function of the mitotic spindle in mammalian cells in vivo. Cell Growth Differ. 11:455–465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous