How many mutant p53 molecules are needed to inactivate a tetramer? - PubMed (original) (raw)
How many mutant p53 molecules are needed to inactivate a tetramer?
Wan Mui Chan et al. Mol Cell Biol. 2004 Apr.
Abstract
The tumor suppressor p53 is transcription factor composed of four identical subunits. The majority of the mutations in p53 are missense mutations that impair DNA binding. On the other hand, the p53-related p63 and p73 genes are rarely mutated, but many cell types express natural variants lacking the N-terminal transactivation domain (NDelta). Compelling evidence indicates that both the DNA binding-defective and NDelta mutants can impair the function of wild-type p53 in a dominant-negative manner. Interestingly, it is uncertain how many mutant subunit(s) a p53 tetramer can tolerate. In this study, we first made theoretical predictions based on the number of mutant p53 monomers needed to inactivate a tetramer and then tested how well the experimental data fit the predicted values. Surprisingly, these experiments reveal that DNA binding-defective p53 mutants (R249S and R273H) are very ineffective in impairing the transcriptional activity of p53: at least three mutants are required to inactivate a tetramer. In marked contrast, p53NDelta is a very potent inhibitor of p53: one NDelta subunit per tetramer is sufficient to abolish the transcriptional activity. DNA binding is not necessary for the NDelta proteins to inactivate p53. Similarly, NDelta variants of p63 and p73 are also powerful inhibitors of members of the p53 family. These results have important implications for our thinking about the mechanism of tumorigenesis involving missense p53 mutants or the N-terminally truncated isoforms.
Figures
FIG. 1.
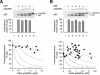
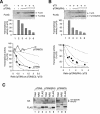
Theoretical prediction of the activity of p53 tetramer. (A) The probabilities of the formation of tetramers with different compositions. W denotes wild-type and M denotes mutant p53. The probabilities (P) of forming different tetramers are given, where the ratio r is the concentration of M/concentration of W. (B) Theoretical prediction of p53 activity with different relative levels of mutant p53. The activity against the ratio of M/W is calculated from the probability of forming the different tetramers shown in panel A. The various curves are based on the assumption that tetramers are only active with the indicated number of wild-type p53 subunits as follows: 4W, 4; 3W, ≥3; 2W, ≥2; or 1W, ≥1. (C) Theoretical prediction of p53 activity for proportional inhibition by a p53 mutant. The activity is calculated according to the assumption that it is directly proportional to the number of wild-type molecules in the tetramer. The four dotted lines are identical to those from panel B for reference. (D) Schematic diagram of the p53 constructs used in this study. The positions of the various structural elements are shown to scale. WT, wild type; LW, point mutation L22Q+W23S.
FIG. 2.
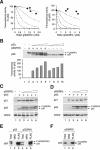
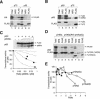
DNA binding-defective p53 mutants inhibit p53 tetramer inefficiently. (A) Inhibition curve of p53(RS) on p53 activity. H1299 cells were transfected with plasmids expressing an MDM2-promoter luciferase reporter and β-galactosidase. Plasmids expressing wild-type p53 and increasing amounts of FLAG-p53(RS) were cotransfected as indicated. Cell extracts were prepared at 24 h after transfection. Both p53 and FLAG-p53(RS) were detected together by immunoblotting with a monoclonal antibody against p53. The luciferase activity was measured, normalized with the β-galactosidase activity to correct for variations in transfection efficiency between samples, and plotted as a percentage of p53 alone (middle panel). The relative expression level of p53 and FLAG-p53(RS) was quantified as described in Materials and Methods and plotted against p53 activity (filled circles, bottom panel). Data from several experiments were pooled. The dotted lines represent the predicted inhibition curves as shown in Fig. 1B. (B) RH mutant. Experiments were performed as described for panel A except that FLAG-p53(RH) was used instead of p53(RS).
FIG. 3.
RS and RH mutants form complexes with p53 without significant effect on its transcriptional activities. (A) At least three p53(RS) or p53(RH) mutants per tetramer are needed to impair the transactivation of p21_CIP1/WAF1_ promoter. Wild-type p53 was coexpressed with various levels of FLAG-tagged p53(RS) or p53(RH) as described in the legend of Fig. 2, except that the p21_CIP1/WAF1_ promoter-luciferase reporter was used instead of the MDM2 promoter. The dotted lines represent the predicted inhibition curves as shown in Fig. 1B. (B) p53(RH) does not inhibit p53 transcriptional activity. Cells were transfected with plasmids expressing an MDM2-promoter luciferase reporter and β-galactosidase. A constant amount of FLAG-p53(RH) and various amounts of wild-type p53 were expressed as indicated. Cell extracts were prepared, and the luciferase and β-galactosidase activities were determined (lower panel). The transcriptional activity was expressed as a percentage of the lowest concentration of p53 used (lane 7). The expression of p53 and FLAG-p53(RH) was detected by immunoblotting. (C) Expression of endogenous p21_CIP1/WAF1_ is only weakly impaired by p53(RS). Cells were transfected with plasmids expressing wild-type p53 and FLAG-p53(RS) as indicated. Cell extracts were prepared at 24 h after transfection, and the expression of p21_CIP1/WAF1_ was detected by immunoblotting. Recombinant p53 and FLAG-p53(RS) were detected together by immunoblotting with a monoclonal antibody against p53. Equal loading of lysates was confirmed by immunoblotting for CDC2. (D) Expression of endogenous p21_CIP1/WAF1_ is only weakly impaired by p53(RH). The experiment was performed as described for panel C except that p53(RH) was used instead ofp53(RS). (E) p53-p53(RH) complexes are formed as efficiently as p53-p53. Untagged p53 was coexpressed with either FLAG-p53 (lanes 1 to 3) or FLAG-p53(RH) (lanes 4 to 6). Cell extracts were prepared and 100 μg was subjected to immunoprecipitation with either control normal rabbit serum (NRS) or FLAG antiserum as indicated. The immunoprecipitates were immunoblotted for p53. Total cell lysates (10 μg) were applied to indicate the inputs. (F) p53(RS) binds efficiently to wild-type p53. Untagged p53 was coexpressed with FLAG-p53(RS). Cell extracts were prepared, immunoprecipitated with NRS or anti-FLAG antiserum, and immunoblotted for p53 as described for panel C.
FIG. 4.
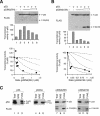
Inhibition of p53 tetramer by transcriptional-incompetent mutants. (A) Inhibition curve of p53NΔ(RH). Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔ(RH), similar to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody against the FLAG epitope. (B) Inhibition curve of p53NΔ159. Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔ159 according to the method described in the legend of Fig. 2. (C) Binding of p53 to different truncation mutants. Untagged p53 was coexpressed with FLAG-tagged p53, p53CΔ, p53NΔ(RH), or p53NΔ159 as indicated. Cell extracts were prepared and subjected to immunoprecipitation followed by immunoblotting for p53 and FLAG according to the method described in the legend of Fig. 3E. NRS, normal rabbit serum.
FIG. 5.
p53 tetramer is strongly inhibited by a mutant lacking the transactivation domain. (A) Inhibition curve of p53NΔ. Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔ according to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody against the FLAG epitope. (B) Association between p53 and p53NΔ. Untagged p53 was coexpressed with FLAG-p53NΔ. Cell extracts were prepared and subjected to immunoprecipitation followed by immunoblotting for p53 and FLAG according to the method described in the legend of Fig. 3E. (C) p53NΔ does not trigger the activation of non-DNA binding p53. Cells were transfected with plasmids expressing an MDM2-promoter luciferase reporter and β-galactosidase. FLAG-tagged p53(RH) was coexpressed with p53NΔ or p53NΔCΔ as indicated. Cell extracts were prepared, and the expression of the recombinant proteins was detected by immunoblotting. The luciferase activity was measured and normalized with the β-galactosidase activity. Lane 12 shows the luciferase activity of wild-type p53 as control. (D) Inhibition curve of p53(LW). Transcriptional assays were performed with p53 and various levels of FLAG-p53(LW) according to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody 421 against p53. NRS, normal rabbit serum.
FIG. 6.
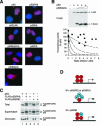
Nuclear localization and chromatin binding of p53 mutants. (A) Localization of p53 mutants. FLAG-tagged p53 or the indicated mutants were transfected into H1299 cells. The cells were fixed, and the localization of the recombinant proteins was analyzed by indirect immunofluorescence microscopy with a monoclonal antibody against FLAG (red). Hoechst 33258 dye was used for nuclear staining (blue). (B) p53NΔCΔ mutant does not inhibit p53 tetramer. Transcriptional assays were performed with FLAG-p53 and various levels of FLAG-p53NΔCΔ according to the method described in the legend of Fig. 2. The recombinant proteins were detected by immunoblotting with a monoclonal antibody against the FLAG epitope. (C) Wild-type p53 triggers binding of RS and RH to chromatin. H1299 cells were transfected with plasmids expressing p53, FLAG-p53(RS), and FLAG-p53(RH) as indicated. The cells were harvested and subjected to chromatin fractionation as described in Materials and Methods. Total cell lysates, supernatant, and chromatin-bound fractions were subjected to immunoblotting for p53. The fractionation was validated by immunoblotting for histone H3 (chromatin bound) and prohibitin (supernatant) (data not shown). (D) Model of inhibition of tetramer by p53 mutants. For DNA binding-defective mutants, at least three mutant molecules per tetramer are required to inactivate transcription. For NΔ mutants, one molecule per tetramer is sufficient to abolish transcription.
FIG. 7.
N-terminally truncated mutants of p73 can inactivate p53. (A) p73NΔ but not p73NΔCΔ inhibits p53. FLAG-tagged p53 was coexpressed with p73NΔ or p73NΔCΔ as indicated. Transcriptional assays were performed according to the method described in the legend of Fig. 2. A summary of several experiments is shown in the bottom panel. (B) Localization of p73 mutants. FLAG-tagged p73 or the indicated mutants were transfected into H1299 cells. The cells were fixed, and the localization of the recombinant proteins was analyzed by indirect immunofluorescence microscopy with a monoclonal antibody against FLAG (red). Hoechst 33258 dye was used for nuclear staining (blue). (C) p73NΔ can form a complex with p53. Untagged p53 was coexpressed with FLAG-p73NΔ. Cell extracts were prepared and subjected to immunoprecipitation followed by immunoblotting for p53 and FLAG according to the method described in the legend of Fig. 3E. (D) The affinity between p53 and p73 is significantly weaker than between p53 and p53. FLAG-p53, HA-p53, and HA-p73 were coexpressed in H1299 cells. Cell extracts were prepared and subjected to immunoprecipitation with normal rabbit serum (NRS) or FLAG antiserum. The immunoprecipitates were immunoblotted for FLAG and HA as indicated. Total cell lysates were applied in lane 1.
FIG. 8.
Inactivation of p73 by p73 lacking the transactivation domain. (A) p73NΔ but not p73NΔCΔ inhibits p73. FLAG-p73 was coexpressed with FLAG-p73NΔ. Transcriptional assays were performed according to the method described in the legend of Fig. 2. A summary of several experiments with p73NΔ as well as p73NΔCΔ is shown in the bottom panel. (B) Inhibition of p73 by p73NΔ(R293H). Transcriptional assays were performed according to the method described in the legend of Fig. 2 to assess the effects of FLAG-p73(RH) on FLAG-p73. (C) p73 binds to p73NΔ but not to p73 lacking the putative tetramerization domain. HA-p73 was coexpressed with FLAG-tagged p73NΔ, p73NΔ(RH), or p73NΔCΔ as indicated. Cell extracts were prepared, subjected to immunoprecipitation with FLAG antiserum, and followed by immunoblotting for HA and FLAG. NRS, normal rabbit serum.
FIG. 9.
A high ratio of p63NΔ or p73NΔ is required for inhibition of p63. (A) p63 binds to p73 but not to p53. HA-p63 was coexpressed with either FLAG-p73 or FLAG-p53 as indicated. Cell extracts were prepared, subjected to immunoprecipitation with normal rabbit serum or FLAG antiserum, and followed by immunoblotting for HA and FLAG. (B) p63NΔ form complexes with p63 and p73. Untagged p63NΔ was coexpressed with either FLAG-p63 or FLAG-p73 as indicated. Cell extracts were prepared, subjected to immunoprecipitation with FLAG antiserum, and followed by immunoblotting for p63 and FLAG. (C) Inhibition of p63 by p63NΔ. Transcriptional assays were performed according to the method described in the legend of Fig. 2 to assess the effects of p63NΔ on FLAG-p63. The recombinant proteins were detected with a monoclonal antibody against p63. (D) p73NΔ can bind to p63. HA-p63 was coexpressed with FLAG-tagged p73NΔ, p73NΔ(RH), or p73NΔCΔ as indicated. Cell extracts were prepared, subjected to immunoprecipitation with normal rabbit serum or FLAG antiserum, and followed by immunoblotting for p63 and FLAG. (E) Inhibition curves of various p73 mutants on p63. Transcriptional assays were performed according to the method described in the legend of Fig. 2 to assess the effects of FLAG-tagged p73NΔ, p73NΔ(RH), or p73NΔCΔ on p63. NRS, normal rabbit serum.
Similar articles
- Functional interplay between MDM2, p63/p73 and mutant p53.
Stindt MH, Muller PA, Ludwig RL, Kehrloesser S, Dötsch V, Vousden KH. Stindt MH, et al. Oncogene. 2015 Aug 13;34(33):4300-10. doi: 10.1038/onc.2014.359. Epub 2014 Nov 24. Oncogene. 2015. PMID: 25417702 Free PMC article. - p21/CDKN1A mediates negative regulation of transcription by p53.
Löhr K, Möritz C, Contente A, Dobbelstein M. Löhr K, et al. J Biol Chem. 2003 Aug 29;278(35):32507-16. doi: 10.1074/jbc.M212517200. Epub 2003 May 13. J Biol Chem. 2003. PMID: 12748190 - Functional inactivation of p73, a homolog of p53 tumor suppressor protein, by human papillomavirus E6 proteins.
Park JS, Kim EJ, Lee JY, Sin HS, Namkoong SE, Um SJ. Park JS, et al. Int J Cancer. 2001 Mar 15;91(6):822-7. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1130>3.0.co;2-0. Int J Cancer. 2001. PMID: 11275986 - The p53 gene family.
Kaelin WG Jr. Kaelin WG Jr. Oncogene. 1999 Dec 13;18(53):7701-5. doi: 10.1038/sj.onc.1202955. Oncogene. 1999. PMID: 10618710 Review. - P53 family: at the crossroads in cancer therapy.
Alsafadi S, Tourpin S, André F, Vassal G, Ahomadegbe JC. Alsafadi S, et al. Curr Med Chem. 2009;16(32):4328-44. doi: 10.2174/092986709789578196. Curr Med Chem. 2009. PMID: 19754415 Review.
Cited by
- Mutant p53: one name, many proteins.
Freed-Pastor WA, Prives C. Freed-Pastor WA, et al. Genes Dev. 2012 Jun 15;26(12):1268-86. doi: 10.1101/gad.190678.112. Genes Dev. 2012. PMID: 22713868 Free PMC article. Review. - Radiosensitization of prostate cancer by priming the wild-type p53-dependent cellular senescence pathway.
Lehmann BD, McCubrey JA, Terrian DM. Lehmann BD, et al. Cancer Biol Ther. 2007 Aug;6(8):1165-70. doi: 10.4161/cbt.6.8.4544. Epub 2007 Aug 5. Cancer Biol Ther. 2007. PMID: 18059157 Free PMC article. Review. - P63 and P73 Activation in Cancers with p53 Mutation.
Cai BH, Hsu YC, Yeh FY, Lin YR, Lu RY, Yu SJ, Shaw JF, Wu MH, Tsai YZ, Lin YC, Bai ZY, Shih YC, Hsu YC, Liao RY, Kuo WH, Hsu CT, Lien CF, Chen CC. Cai BH, et al. Biomedicines. 2022 Jun 23;10(7):1490. doi: 10.3390/biomedicines10071490. Biomedicines. 2022. PMID: 35884795 Free PMC article. Review. - Limited importance of the dominant-negative effect of TP53 missense mutations.
Stoczynska-Fidelus E, Szybka M, Piaskowski S, Bienkowski M, Hulas-Bigoszewska K, Banaszczyk M, Zawlik I, Jesionek-Kupnicka D, Kordek R, Liberski PP, Rieske P. Stoczynska-Fidelus E, et al. BMC Cancer. 2011 Jun 13;11:243. doi: 10.1186/1471-2407-11-243. BMC Cancer. 2011. PMID: 21668955 Free PMC article. - Allele-specific silencing of mutant p53 attenuates dominant-negative and gain-of-function activities.
Iyer SV, Parrales A, Begani P, Narkar A, Adhikari AS, Martinez LA, Iwakuma T. Iyer SV, et al. Oncotarget. 2016 Feb 2;7(5):5401-15. doi: 10.18632/oncotarget.6634. Oncotarget. 2016. PMID: 26700961 Free PMC article.
References
- Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl. 1991. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- Bodner, S. M., J. D. Minna, S. M. Jensen, D. D'Amico, D. Carbone, T. Mitsudomi, J. Fedorko, D. L. Buchhagen, M. M. Nau, A. F. Gazdar, et al. 1992. Expression of mutant p53 proteins in lung cancer correlates with the class of p53 gene mutation. Oncogene 7:743-749. - PubMed
- Cho, Y., S. Gorina, P. D. Jeffrey, and N. P. Pavletich. 1994. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265:346-355. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous