The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development - PubMed (original) (raw)
. 2004 May 15;18(10):1187-97.
doi: 10.1101/gad.1201404. Epub 2004 May 6.
Affiliations
- PMID: 15131082
- PMCID: PMC415643
- DOI: 10.1101/gad.1201404
The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development
Hervé Vaucheret et al. Genes Dev. 2004.
Abstract
MicroRNAs (miRNAs) are endogenous 21-24-nt RNAs that can down-regulate gene expression by pairing to the messages of protein-coding genes to specify mRNA cleavage or repression of productive translation. They act within the RNA-induced silencing complex (RISC), which in animals contains a member of the Argonaute family of proteins. In the present study, we show that Arabidopsis ago1 mutants have increased accumulation of mRNAs known to be targeted for cleavage by miRNAs. In hypomorphic ago1 alleles, this compromised miRNA function occurs without a substantial change in miRNA accumulation, whereas in null alleles it is accompanied by a drop in some of the miRNAs. Therefore, AGO1 acts within the Arabidopsis miRNA pathway, probably within the miRNA-programmed RISC, such that the absence of AGO1 destabilizes some of the miRNAs. We also show that targeting of AGO1 mRNA by miR168 is needed for proper plant development, illustrating the importance of feedback control by this miRNA. Transgenic plants expressing a mutant AGO1 mRNA with decreased complementarity to miR168 overaccumulate AGO1 mRNA and exhibit developmental defects partially overlapping with those of dcl1, hen1, and hyl1 mutants showing a decrease in miRNA accumulation. miRNA targets overaccumulate in miR168-resistant plants, suggesting that a large excess of AGO1 protein interferes with the function of RISC or sequesters miRNAs or other RISC components. Developmental defects induced by a miR168-resistant AGO1 mRNA can be rescued by a compensatory miRNA that is complementary to the mutant AGO1 mRNA, proving the regulatory relationship between miR168 and its target and opening the way for engineering artificial miRNAs in plants.
Figures
Figure 1.
ago1, hen1, and hyl1 mutants exhibit overlapping developmental defects. (A) Rosettes of plants grown under short-day conditions. (B) Flowers of plants grown under long-day conditions.
Figure 2.
ago1, hen1, and hyl1 mutants have increased steady-state levels of miRNA targets. RNA extracted from rosettes of isogenic wild-type or mutant siblings deriving from heterozygote parents and of untransformed plants or 2m-AGO1 transformants was quantified for the indicated mRNA by real-time quantitative PCR using primers surrounding the cleavage site. GAPDH and eEF-1(A4) were used as nontarget controls. Quantifications were normalized to that of ACTIN2, then to the value of the wild-type plants or wild-type siblings, which was arbitrarily fixed to 1.
Figure 3.
miRNA accumulation in ago1 mutants. miRNA accumulation was determined by RNA gel blot analysis using 30 μg (A) or 10μg (B) of the same RNA used for RT–qPCR analyses. Blots were successively hybridized to different probes complementary to miRNAs. (A) miRNA accumulation in the ago1-26 and ago1-27 hypomorphic alleles. (B) miRNA accumulation in the _ago1-3_-null allele.
Figure 4.
Silent mutations in the miR168 complementary site of the AGO1 mRNA induce developmental defects. (A) The WT-AGO1 mRNA naturally contains three mismatches with miR168 (in blue), including a G:U wobble pair. Silent mutations in 2m-AGO1 and 4m-AGO1 constructs introduce two and four additional mismatches (in red), reducing complementarity with miR168. ΔΔ_G_ was calculated using mfold. (B) Representative sets of transformants carrying the WT-AGO1 or 2m-AGO1 construct. (C) Proportion of transformants showing a wild-type phenotype (open bar), an ago1 phenotype caused by late cosuppression (yellow bar), or an mir-AGO1 phenotype caused by AGO1 overexpression (red bar). Plants were transformed with either an empty vector (EV) or the WT-AGO1, 2m-AGO1, or 4m-AGO1 constructs. The number of transformants analyzed is indicated in parentheses. (D) AGO1 mRNA accumulation determined by real-time quantitative PCR in untransformed plants (Col) or plants transformed with the WT-AGO1 or 2m-AGO1 constructs. Quantifications were normalized to that of ACTIN2. The value in Col was arbitrarily fixed to 1. Numbers (#) correspond to the plants shown in B.
Figure 5.
Developmental defects in 2m-AGO1 transformants. (A, top row) Wild-type plant (Col) and dcl1, hen1, and hyl1 mutants. (Middle row) Wild-type plant (Col) and 2m-AGO1 transformants exhibiting curled leaves resembling those of hen1 and hyl1 mutants, at 10 d. Transformants with aberrant cotyledons were occasionally observed (right). (Bottom row) Wild-type plant (Col) and 4m-AGO1 transformants exhibiting a variety of developmental defects, including asymmetric rosette leaf formation and curled or twisted leaves, at 20 d. (B, top row) Wild-type plant (Col) and dcl1 and hyl1 mutants. (Bottom row) Adult wild-type plant (Col) and a representative 2m-AGO1 transformant exhibiting spoon-shaped or twisted anthocyaned leaves resembling those of dcl1 and hyl1 mutants. (C, top row) Inflorescence of a wild-type plant (Col) and of a representative 2m-AGO1 transformant. (Bottom row) Stems and siliques (seed pots) of the same plants. The wild-type Col plant is fertile, whereas the 2m-AGO1 transformant is sterile with aborted siliques.
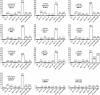
Figure 6.
Compensatory mutations in the MIR168a gene rescue developmental defects induced by silent mutations in the miR168 complementary site of the AGO1 mRNA. (A) The MIR168a gene encodes a primary transcript that is partially paired (unpaired nucleotides are purple). The miRNA is boxed. Compensatory mutations in the 4m-MIR168a transgene (red) conserved the structure of the primary transcript and restored pairing with the 4m-AGO1 mRNA. Original mismatches (blue) were kept. ΔΔ_G_ was calculated using mfold. (B) Representative sets of transformants carrying the 4m-AGO1 construct alone or the 4m-AGO1 and 4m-MIR168a constructs together. (C) Proportion of transformants showing a wild-type phenotype (open bar), an ago1 phenotype caused by late cosuppression (yellow bar), or a mir-AGO1 phenotype caused by AGO1 overexpression (red bar). Plants were transformed with the 4m-AGO1 or 4m-MIR168a constructs or both. The number of transformants analyzed is indicated in parentheses. (D) Accumulation of the compensatory miRNA (m4-miR168) in double transformants carrying the 4m-AGO1 and 4m-MIR168a constructs. RNA gel blot analysis was performed using 20 μg of total RNA extracted from two nontransformed plants (Col) and eight independent double transformants. The blot was hybridized with a probe complementary to 4m-miR168, stripped, rehybridized with a probe complementary to miR168, stripped, and finally rehybridized with the two probes simultaneously.
Similar articles
- AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1.
Vaucheret H, Mallory AC, Bartel DP. Vaucheret H, et al. Mol Cell. 2006 Apr 7;22(1):129-36. doi: 10.1016/j.molcel.2006.03.011. Mol Cell. 2006. PMID: 16600876 Free PMC article. - Controlled RISC loading efficiency of miR168 defined by miRNA duplex structure adjusts ARGONAUTE1 homeostasis.
Dalmadi Á, Miloro F, Bálint J, Várallyay É, Havelda Z. Dalmadi Á, et al. Nucleic Acids Res. 2021 Dec 16;49(22):12912-12928. doi: 10.1093/nar/gkab1138. Nucleic Acids Res. 2021. PMID: 34850097 Free PMC article. - MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region.
Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. Mallory AC, et al. EMBO J. 2004 Aug 18;23(16):3356-64. doi: 10.1038/sj.emboj.7600340. Epub 2004 Jul 29. EMBO J. 2004. PMID: 15282547 Free PMC article. - Plant and animal microRNAs: similarities and differences.
Millar AA, Waterhouse PM. Millar AA, et al. Funct Integr Genomics. 2005 Jul;5(3):129-35. doi: 10.1007/s10142-005-0145-2. Epub 2005 May 5. Funct Integr Genomics. 2005. PMID: 15875226 Review. - MicroRNAs in disease and potential therapeutic applications.
Soifer HS, Rossi JJ, Saetrom P. Soifer HS, et al. Mol Ther. 2007 Dec;15(12):2070-9. doi: 10.1038/sj.mt.6300311. Epub 2007 Sep 18. Mol Ther. 2007. PMID: 17878899 Review.
Cited by
- Lessons on RNA silencing mechanisms in plants from eukaryotic argonaute structures.
Poulsen C, Vaucheret H, Brodersen P. Poulsen C, et al. Plant Cell. 2013 Jan;25(1):22-37. doi: 10.1105/tpc.112.105643. Epub 2013 Jan 9. Plant Cell. 2013. PMID: 23303917 Free PMC article. - Developmentally regulated expression and complex processing of barley pri-microRNAs.
Kruszka K, Pacak A, Swida-Barteczka A, Stefaniak AK, Kaja E, Sierocka I, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z. Kruszka K, et al. BMC Genomics. 2013 Jan 16;14:34. doi: 10.1186/1471-2164-14-34. BMC Genomics. 2013. PMID: 23324356 Free PMC article. - Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs.
McCue AD, Nuthikattu S, Slotkin RK. McCue AD, et al. RNA Biol. 2013 Aug;10(8):1379-95. doi: 10.4161/rna.25555. Epub 2013 Jul 2. RNA Biol. 2013. PMID: 23863322 Free PMC article. - Identification of miRNAs in sorghum by using bioinformatics approach.
Katiyar A, Smita S, Chinnusamy V, Pandey DM, Bansal K. Katiyar A, et al. Plant Signal Behav. 2012 Feb;7(2):246-59. doi: 10.4161/psb.18914. Epub 2012 Feb 1. Plant Signal Behav. 2012. PMID: 22415044 Free PMC article. - Exploring microRNA Signatures of DNA Damage Response Using an Innovative System of Genotoxic Stress in Medicago truncatula Seedlings.
Gualtieri C, Gianella M, Pagano A, Cadeddu T, Araújo S, Balestrazzi A, Macovei A. Gualtieri C, et al. Front Plant Sci. 2021 Mar 9;12:645323. doi: 10.3389/fpls.2021.645323. eCollection 2021. Front Plant Sci. 2021. PMID: 33767724 Free PMC article.
References
- Ambros V. 2003. MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 113: 673–676. - PubMed
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297. - PubMed
- Bechtold N. and Pelletier, G. 1998. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82: 259–266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials