Absence of the TAP2 human recombination hotspot in chimpanzees - PubMed (original) (raw)
Comparative Study
Absence of the TAP2 human recombination hotspot in chimpanzees
Susan E Ptak et al. PLoS Biol. 2004 Jun.
Abstract
Recent experiments using sperm typing have demonstrated that, in several regions of the human genome, recombination does not occur uniformly but instead is concentrated in "hotspots" of 1-2 kb. Moreover, the crossover asymmetry observed in a subset of these has led to the suggestion that hotspots may be short-lived on an evolutionary time scale. To test this possibility, we focused on a region known to contain a recombination hotspot in humans, TAP2, and asked whether chimpanzees, the closest living evolutionary relatives of humans, harbor a hotspot in a similar location. Specifically, we used a new statistical approach to estimate recombination rate variation from patterns of linkage disequilibrium in a sample of 24 western chimpanzees (Pan troglodytes verus). This method has been shown to produce reliable results on simulated data and on human data from the TAP2 region. Strikingly, however, it finds very little support for recombination rate variation at TAP2 in the western chimpanzee data. Moreover, simulations suggest that there should be stronger support if there were a hotspot similar to the one characterized in humans. Thus, it appears that the human TAP2 recombination hotspot is not shared by western chimpanzees. These findings demonstrate that fine-scale recombination rates can change between very closely related species and raise the possibility that rates differ among human populations, with important implications for linkage-disequilibrium based association studies.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
Figure 1. Patterns of Pairwise LD in Humans and Chimpanzees
Only SNPs with minor allele frequencies above 0.1 are included. The rows correspond to the consecutive SNPs in the region, as do the columns. Each cell indicates the extent of LD between a pair of sites, as measured by |_D_′| (estimated using the Expectation Maximization algorithm, as implemented by Arlequin:
).
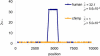
Figure 2. Estimates of the Recombination Hotspot Intensity, λ, Based on Genotype Data
We assumed that, if the hotspot is present, it is in the same location as estimated by sperm typing in humans (see Materials and Methods). A λ value of one corresponds to the absence of recombination rate variation, while values of λ greater than one indicate a hotspot. The estimates for humans from the UK are shown in blue and those for western chimpanzees in orange.
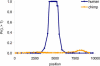
Figure 3. Estimates of Recombination Rate Variation in Humans and Western Chimpanzees
In this model, there is at most one hotspot in the region, the location and width of which are unknown and estimated along with λ and ρ. On the _y_-axis is an estimate of the posterior probability of elevated recombination, Pr(λ > 1), between each pair of consecutive SNPs (plotted at the midpoint position).
Figure 4. Distribution of Variable Sites in the Genomic Region
The positions of sites that differ between humans and chimpanzees are shown on the first line, while the positions of sites polymorphic in humans from the UK or in western chimpanzees are shown on the next two lines. The human hotspot region is underlined. The dashed lines indicate regions not surveyed for variation in western chimpanzees (see Materials and Methods).
Similar articles
- High resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot.
Jeffreys AJ, Ritchie A, Neumann R. Jeffreys AJ, et al. Hum Mol Genet. 2000 Mar 22;9(5):725-33. doi: 10.1093/hmg/9.5.725. Hum Mol Genet. 2000. PMID: 10749979 - Fine-scale recombination patterns differ between chimpanzees and humans.
Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Pääbo S. Ptak SE, et al. Nat Genet. 2005 Apr;37(4):429-34. doi: 10.1038/ng1529. Epub 2005 Feb 18. Nat Genet. 2005. PMID: 15723063 - Evidence for a complex demographic history of chimpanzees.
Fischer A, Wiebe V, Pääbo S, Przeworski M. Fischer A, et al. Mol Biol Evol. 2004 May;21(5):799-808. doi: 10.1093/molbev/msh083. Epub 2004 Feb 12. Mol Biol Evol. 2004. PMID: 14963091 - Meiotic recombination hot spots and human DNA diversity.
Jeffreys AJ, Holloway JK, Kauppi L, May CA, Neumann R, Slingsby MT, Webb AJ. Jeffreys AJ, et al. Philos Trans R Soc Lond B Biol Sci. 2004 Jan 29;359(1441):141-52. doi: 10.1098/rstb.2003.1372. Philos Trans R Soc Lond B Biol Sci. 2004. PMID: 15065666 Free PMC article. Review. - What's so hot about recombination hotspots?
Hey J. Hey J. PLoS Biol. 2004 Jun;2(6):e190. doi: 10.1371/journal.pbio.0020190. Epub 2004 Jun 15. PLoS Biol. 2004. PMID: 15208734 Free PMC article. Review.
Cited by
- Detecting Recombination Hotspots from Patterns of Linkage Disequilibrium.
Wall JD, Stevison LS. Wall JD, et al. G3 (Bethesda). 2016 Aug 9;6(8):2265-71. doi: 10.1534/g3.116.029587. G3 (Bethesda). 2016. PMID: 27226166 Free PMC article. - Combining sperm typing and linkage disequilibrium analyses reveals differences in selective pressures or recombination rates across human populations.
Clark VJ, Ptak SE, Tiemann I, Qian Y, Coop G, Stone AC, Przeworski M, Arnheim N, Di Rienzo A. Clark VJ, et al. Genetics. 2007 Feb;175(2):795-804. doi: 10.1534/genetics.106.064964. Epub 2006 Dec 6. Genetics. 2007. PMID: 17151245 Free PMC article. - Parallel Molecular Evolution in Pathways, Genes, and Sites in High-Elevation Hummingbirds Revealed by Comparative Transcriptomics.
Lim MCW, Witt CC, Graham CH, Dávalos LM. Lim MCW, et al. Genome Biol Evol. 2019 Jun 1;11(6):1552-1572. doi: 10.1093/gbe/evz101. Genome Biol Evol. 2019. PMID: 31114863 Free PMC article. - Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes.
Muñoz-Fuentes V, Di Rienzo A, Vilà C. Muñoz-Fuentes V, et al. PLoS One. 2011;6(11):e25498. doi: 10.1371/journal.pone.0025498. Epub 2011 Nov 10. PLoS One. 2011. PMID: 22102853 Free PMC article. - The Genetic Architecture of Natural Variation in Recombination Rate in Drosophila melanogaster.
Hunter CM, Huang W, Mackay TF, Singh ND. Hunter CM, et al. PLoS Genet. 2016 Apr 1;12(4):e1005951. doi: 10.1371/journal.pgen.1005951. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27035832 Free PMC article.
References
- Ashburner M. Cold Spring Harbor (New York): Cold Spring Harbor Laboratory Press; 1989. Drosophila: A laboratory handbook; 1331 pp.
- Badge RM, Yardley J, Jeffreys AJ, Armour JA. Crossover breakpoint mapping identifies a subtelomeric hotspot for male meiotic recombination. Hum Mol Genet. 2000;9:1239–1244. - PubMed
- Beck S, Abdulla S, Alderton RP, Glynne RJ, Gut IG, et al. Evolutionary dynamics of non-coding sequences within the class II region of the human MHC. J Mol Biol. 1996;255:1–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials