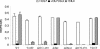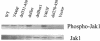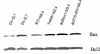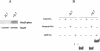Distinct regions of the interleukin-7 receptor regulate different Bcl2 family members - PubMed (original) (raw)
Distinct regions of the interleukin-7 receptor regulate different Bcl2 family members
Qiong Jiang et al. Mol Cell Biol. 2004 Jul.
Abstract
The antiapoptotic function of the interleukin-7 (IL-7) receptor is related to regulation of three members of the Bcl2 family: synthesis of Bcl2, phosphorylation of Bad, and cytosolic retention of Bax. Here we show that, in an IL-7-dependent murine T-cell line, different regions of the IL-7 receptor initiate the signal transduction pathways that regulate these proteins. Both Box1 and Y449 are required to signal Bcl2 synthesis and Bax cytosolic retention. This suggests a sequential model in which Jak1, which binds to Box1, is first activated and then phosphorylates Y449, leading to Bcl2 and Bax regulation, accounting for approximately 90% of the survival function. Phosphorylation of Bad required Box1 but not Y449, suggesting that Jak1 also initiates an additional signaling cascade that accounts for approximately 10% of the survival function. Stat5 was activated from the Y449 site but only partially accounted for the survival signal. Proliferation required both Y449 and Box1. Thymocyte development in vivo showed that deletion of Y449 eliminated 90% of alphabeta T-cell development and completely eliminated gammadelta T-cell development, whereas deleting Box 1 completely eliminated both alphabeta and gammadelta T-cell development. Thus the IL-7 receptor controls at least two distinct pathways, in addition to Stat5, that are required for cell survival.
Figures
FIG. 1.
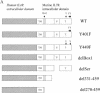
Construction of chimeric IL-7 receptors and expression on D1 cells. (A) Structures of the chimeric mutant receptors used in this study. Constructs contain the hIL-4R extracellular domain, mIL-7R transmembrane domain (TM), and mIL-7R intracellular region. S, serine-rich domain; T, tyrosine-containing domain. (B) Structure of the pMIG vector used to generate retroviruses. LTR, long terminal repeat; IRES, internal ribosomal entry site; MCS, multiple cloning site. (C) Flow-cytometric analysis of D1 cells infected with the parental retrovirus (Vector) or retroviruses. Cells were sorted for GFP and then stained for surface expression of hIL-4R.
FIG. 1.
Construction of chimeric IL-7 receptors and expression on D1 cells. (A) Structures of the chimeric mutant receptors used in this study. Constructs contain the hIL-4R extracellular domain, mIL-7R transmembrane domain (TM), and mIL-7R intracellular region. S, serine-rich domain; T, tyrosine-containing domain. (B) Structure of the pMIG vector used to generate retroviruses. LTR, long terminal repeat; IRES, internal ribosomal entry site; MCS, multiple cloning site. (C) Flow-cytometric analysis of D1 cells infected with the parental retrovirus (Vector) or retroviruses. Cells were sorted for GFP and then stained for surface expression of hIL-4R.
FIG. 1.
Construction of chimeric IL-7 receptors and expression on D1 cells. (A) Structures of the chimeric mutant receptors used in this study. Constructs contain the hIL-4R extracellular domain, mIL-7R transmembrane domain (TM), and mIL-7R intracellular region. S, serine-rich domain; T, tyrosine-containing domain. (B) Structure of the pMIG vector used to generate retroviruses. LTR, long terminal repeat; IRES, internal ribosomal entry site; MCS, multiple cloning site. (C) Flow-cytometric analysis of D1 cells infected with the parental retrovirus (Vector) or retroviruses. Cells were sorted for GFP and then stained for surface expression of hIL-4R.
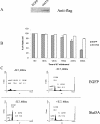
FIG. 2.
MTT analysis of the response of cells to various mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and cultured for 48 h with 50 ng of mIL-7/ml or 50 ng of hIL-4/ml. Results are from one representative experiment of three performed. Chimeric constructs imparted a response to hIL-4. Deletion of Box1 or mutation of Y449 in the intracellular domain of IL-7R severely impaired the response. OD(570-620), absorbance at 570 nm minus background absorbance at 620 nm.
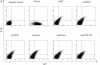
FIG. 3.
Cell cycle profile of the response of cells to various mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and cultured for 72 h in the presence of 50 ng of hIL-4/ml and then stained with PI, followed by flow-cytometric analysis. Results are from one representative experiment of two performed. Percentages of G1/G0, S, G2/M, and sub-G1 cells are shown in the figure. Deletion of Box1 resulted in rapid apoptosis, whereas Y449 mutation induced G1 cell cycle arrest followed by apoptosis.
FIG. 4.
Jak1 activation versus mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and stimulated with 50 ng of hIL-4/ml for 15 min. Total Triton X-100 cell lysates were resolved by sodium dodecyl sulfate-PAGE, and Western blotting was performed with an antibody against phospho-Jak1 or Jak1. The activation of Jak1 required Box1 but not Y449.
FIG. 5.
bcl2 mRNA induction versus mutations and deletions in the intracellular domain of IL-7R. Transfected or nontransfected D1 cells were maintained in mIL-7, washed, and stimulated with either mIL-7 or hIL-4 for 4 h. Total RNA was extracted, and bcl2 transcripts were analyzed by RPA. Induction of bcl2 required both Box1 and Y449. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIG. 6.
Bax cytosolic retention versus mutations and deletions in the intracellular domain of IL-7R. Transfected or nontransfected D1 cells were maintained in mIL-7, washed, and stimulated with either mIL-7 or hIL-4 for 8 h. Cells were disrupted, and mitochondrial protein fractions were prepared and immunoblotted with an anti-Bax or anti-Bcl2 antibody. Cytosolic retention of Bax required both Box1 and Y449.
FIG. 7.
Bad phosphorylation versus mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were maintained in mIL-7, washed, and stimulated with hIL-4 for 4 h. Total Triton X-100 cell lysates were analyzed by Western blotting with an antibody against phospho-Bad or Bad. Phosphorylation of Bad required Box1 but not Y449.
FIG. 8.
Stat activation versus mutations and deletions of the intracellular domain of IL-7R. Transfected D1 cells were deprived of IL-7 overnight and then stimulated with hIL-4 (+) or without hIL-4 (−) for 10 min. Total Triton X-100 cell lysates were analyzed by Western blotting with antibodies against phospho-Stat1, phospho-Stat3, phospho-Stat5, or Stat5. Phosphorylation of Stat5 required both Box 1 and Y449. Phosphorylation of Stat1 and -3 required Box1 but not Y449.
FIG. 9.
Activation of Stat5 in D1 cells following IL-7 stimulation. (A) D1 cells were deprived of IL-7 overnight and then stimulated with mIL-7 for 10 min. (B) D1 cells were deprived of IL-7 overnight and then stimulated with mIL-7 (50 ng/ml) for 2 h. Nuclear extracts were analyzed by EMSA using a labeled Stat5 consensus oligonucleotide. Specificity was evaluated by competition with cold DNA or with an antibody (Ab) against Stat5. Results show that IL-7 induced Stat5 nuclear translocation and DNA binding.
FIG. 10.
Survival effect of expression of active Stat5A on D1 cells following IL-7 withdrawal. (A) Nuclear proteins were prepared from D1 cells infected with parental retrovirus (EGFP) or with retrovirus harboring active Stat5A (stat5A) and subjected to Western blotting with a Flag antibody. (B) EGFP- or Stat5A-expressing cells were washed twice with PBS and cultured without mIL-7 for the indicated times. Percentages of viable cells were determined by PI staining. (C) EGFP (top)- or Stat5A (bottom)-expressing infected D1 cells were cultured with or without mIL-7 (50 ng/ml) for 48 h and stained with PI following flow-cytometric analysis. Percentages of G1/G0, S, G2/M.and sub-G1 cells are shown. Results show that activated Stat5 delayed death of D1 cells deprived of IL-7 but did not induce cell division.
FIG. 11.
Active Stat5A versus bcl2 expression following IL-7 withdrawal. D1 cells infected with control EGFP or Stat5A retrovirus supernatant were deprived of IL-7 for the indicated times. (A) Total RNA was prepared and subjected to RPA for expression of bcl2 or GAPDH as a loading control. As positive controls, cells were stimulated with mIL-7 for 4 h after overnight starvation (+4hr) or were continuously cultured in the presence of mIL-7 (N). (B) Total Triton X-100 extracts were prepared. Western blotting was performed with either a Bcl2 or β-actin antibody. Results show that activated Stat5 was insufficient to maintain high expression of Bcl2 in the absence of IL-7. However, activated Stat5 increased the expression of bcl2 in the presence of IL-7 and sustained bcl2 expression for 4 h following IL-7 withdrawal. Bcl2 protein levels have a long half-life in D1 cells, and activated Stat5 extended the levels of Bcl2 following IL-7 withdrawal.
FIG. 12.
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
FIG. 12.
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
FIG. 12.
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
FIG. 12.
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
FIG. 12.
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
FIG. 12.
Reconstitution of IL-7R−/− T-cell development in vivo. mIL-7R (rather than chimeric) constructs (and GFP) were tested for expression by introducing them into hematopoietic cell lines C1498 and BW5147 by retrovirus infection. Twenty-four hours later, surface expression of mIL-7R (together with GFP expression) was analyzed by fluorescence-activated cell sorting. (A) GFP versus surface expression of mIL-7R in infected C1498 cells. (B) GFP versus surface expression of mIL-7R in infected BW5147 cells. Receptor constructs (and GFP) were introduced by retroviral transfer into IL-7R−/− hematopoietic stem cells, which were injected into Rag1−/− recipients. One month later, thymocytes and spleen cells were analyzed by flow microfluorimetry, gating on GFP+ cells. (C) Transfection efficiency in bone marrow stem cells. Forty-eight hours after retroviral infection, hematopoietic stem cells were analyzed for expression of GFP. Percentages of GFP-positive cells are indicated and are similar for different receptor constructs. (D) Numbers of GFP-positive cells in different groups were obtained based on percentages of GFP-positive cells and total cells in each thymus. Data are from three experiments using six individual mice. (E) Thymocytes were stained for CD4 and CD8. The numbers shown indicate the number (106) of cells of each subset per thymus. Deleting Box1 completely eliminated thymocyte development, whereas Y449F eliminated approximately 90%. There was no effect of deleting the serine region or Y401F. (F) Spleen cells were stained for αβ and γδ T cells. Percentages of γδ T cell are shown. mY449F completely eliminated γδ T-cell development, as did deleting Box1 or aa 270 to 459. No effect was seen for mY401F or deletion of the serine region.
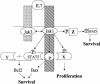
FIG. 13.
Current model for IL-7R regulation of survival and proliferation. IL-7 binding to the receptor brings together the α and γc chains and their associated kinases, Jak1 (bound to Box1) and Jak3, respectively. The two kinases phosphorylate one another and increase their enzymatic activity. These kinases then phosphorylate Y449, which serves as a docking site for Stat5 and additional unknown proteins that are required for synthesis of Bcl2, cytosolic retention of Bax, and cell proliferation. A second pathway leads from Jak1 (independent of Y449) and induces phosphorylation of Bad, Stat1, and Stat3.
Similar articles
- Activation of STAT5 by IL-4 relies on Janus kinase function but not on receptor tyrosine phosphorylation, and can contribute to both cell proliferation and gene regulation.
Friedrich K, Kammer W, Erhardt I, Brändlein S, Sebald W, Moriggl R. Friedrich K, et al. Int Immunol. 1999 Aug;11(8):1283-94. doi: 10.1093/intimm/11.8.1283. Int Immunol. 1999. PMID: 10421786 - Cooperation between STAT5 and phosphatidylinositol 3-kinase in the IL-3-dependent survival of a bone marrow derived cell line.
Rosa Santos SC, Dumon S, Mayeux P, Gisselbrecht S, Gouilleux F. Rosa Santos SC, et al. Oncogene. 2000 Feb 24;19(9):1164-72. doi: 10.1038/sj.onc.1203418. Oncogene. 2000. PMID: 10713704 - Signaling via the IL-2 and IL-7 receptors from the membrane to the nucleus.
Leonard WJ, Imada K, Nakajima H, Puel A, Soldaini E, John S. Leonard WJ, et al. Cold Spring Harb Symp Quant Biol. 1999;64:417-24. doi: 10.1101/sqb.1999.64.417. Cold Spring Harb Symp Quant Biol. 1999. PMID: 11232316 Review. No abstract available. - Regulation of Bcl2 phosphorylation and potential significance for leukemic cell chemoresistance.
Deng X, Kornblau SM, Ruvolo PP, May WS Jr. Deng X, et al. J Natl Cancer Inst Monogr. 2001;(28):30-7. doi: 10.1093/oxfordjournals.jncimonographs.a024254. J Natl Cancer Inst Monogr. 2001. PMID: 11158204 Review.
Cited by
- Incorporating IL7 receptor alpha signaling in the endodomain of B7H3-targeting chimeric antigen receptor T cells mediates antitumor activity in glioblastoma.
Sakunrangsit N, Khuisangeam N, Inthanachai T, Yodsurang V, Taechawattananant P, Suppipat K, Tawinwung S. Sakunrangsit N, et al. Cancer Immunol Immunother. 2024 Apr 15;73(6):98. doi: 10.1007/s00262-024-03685-7. Cancer Immunol Immunother. 2024. PMID: 38619641 Free PMC article. - DIALing-up the preclinical characterization of gene-modified adoptive cellular immunotherapies.
Giardino Torchia ML, Moody G. Giardino Torchia ML, et al. Front Immunol. 2023 Nov 28;14:1264882. doi: 10.3389/fimmu.2023.1264882. eCollection 2023. Front Immunol. 2023. PMID: 38090585 Free PMC article. Review. - IL-7 receptor signaling drives human B-cell progenitor differentiation and expansion.
Kaiser FMP, Janowska I, Menafra R, de Gier M, Korzhenevich J, Pico-Knijnenburg I, Khatri I, Schulz A, Kuijpers TW, Lankester AC, Konstantinidis L, Erlacher M, Kloet S, van Schouwenburg PA, Rizzi M, van der Burg M. Kaiser FMP, et al. Blood. 2023 Sep 28;142(13):1113-1130. doi: 10.1182/blood.2023019721. Blood. 2023. PMID: 37369082 Free PMC article. - Beyond Corticoresistance, A Paradoxical Corticosensitivity Induced by Corticosteroid Therapy in Pediatric Acute Lymphoblastic Leukemias.
Angot L, Schneider P, Vannier JP, Abdoul-Azize S. Angot L, et al. Cancers (Basel). 2023 May 18;15(10):2812. doi: 10.3390/cancers15102812. Cancers (Basel). 2023. PMID: 37345151 Free PMC article. Review. - High glucose-induced IL-7/IL-7R upregulation of dermal fibroblasts inhibits angiogenesis in a paracrine way in delayed diabetic wound healing.
Gao R, Zhou P, Li Y, Li Q. Gao R, et al. J Cell Commun Signal. 2023 Sep;17(3):1023-1038. doi: 10.1007/s12079-023-00754-x. Epub 2023 May 22. J Cell Commun Signal. 2023. PMID: 37217704 Free PMC article.
References
- Akashi, K., M. Kondo, U. von Freeden-Jeffry, R. Murray, and I. L. Weissman. 1997. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell 89:1033-1041. - PubMed
- Benbernou, N., K. Muegge, and S. K. Durum. 2000. Interleukin (IL)-7 induces rapid activation of Pyk2, which is bound to Janus kinase 1 and IL-7Rα. J. Biol. Chem. 275:7060-7065. - PubMed
- Cao, X., E. W. Shores, L. J. Hu, M. R. Anver, B. L. Kelsall, S. M. Russell, J. Drago, M. Noguchi, A. Grinberg, E. T. Bloom, et al. 1995. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 2:223-238. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous