Muscleblind proteins regulate alternative splicing - PubMed (original) (raw)
Muscleblind proteins regulate alternative splicing
Thai H Ho et al. EMBO J. 2004.
Abstract
Although the muscleblind (MBNL) protein family has been implicated in myotonic dystrophy (DM), a specific function for these proteins has not been reported. A key feature of the RNA-mediated pathogenesis model for DM is the disrupted splicing of specific pre-mRNA targets. Here we demonstrate that MBNL proteins regulate alternative splicing of two pre-mRNAs that are misregulated in DM, cardiac troponin T (cTNT) and insulin receptor (IR). Alternative cTNT and IR exons are also regulated by CELF proteins, which were previously implicated in DM pathogenesis. MBNL proteins promote opposite splicing patterns for cTNT and IR alternative exons, both of which are antagonized by CELF proteins. CELF- and MBNL-binding sites are distinct and regulation by MBNL does not require the CELF-binding site. The results are consistent with a mechanism for DM pathogenesis in which expanded repeats cause a loss of MBNL and/or gain of CELF activities, leading to misregulation of alternative splicing of specific pre-mRNA targets.
Figures
Figure 1
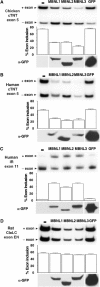
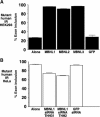
MBNL1, 2 and 3 regulate splicing of cTNT and IR alternative exons. Human and chicken cTNT and human IR minigenes were expressed with or without each of the three GFP–MBNL fusion proteins or with GFP alone. Duplicate transfections were used for extraction of RNA and protein. Inclusion of cTNT exon 5 or IR exon 11 was assayed by RT–PCR. Percent exon inclusion is calculated as ((mRNA+exon)/(mRNA−exon+mRNA+exon)) × 100. Results are derived from at least three independent experiments. Expression of GFP–MBNL1 (∼72 kDa), GFP–MBNL2 (∼58 kDa), GFP–MBNL3 (∼70 kDa) and EGFP (∼27 kDa) was detected by Western blot analysis using an anti-GFP monoclonal antibody. All three MBNL proteins promote exon 5 skipping of (A) chicken and (B) human cTNT exon 5 in primary skeletal muscle cultures. (C) All three MBNL proteins promote exon 11 inclusion in a human IR minigene in HEK293 cells. (D) MBNL proteins have minimal effects on splicing of exon EN in a clathrin light-chain B minigene in primary skeletal muscle cultures.
Figure 2
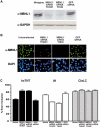
Endogenous MBNL1 regulates the splicing of human cTNT and IR minigenes. siRNA and minigenes were transfected into HeLa cells. (A) Western blot confirming depletion of endogenous MBNL1 by independent transfection of two different siRNA constructs using the MBNL1 monoclonal (mAb) 3A4, which recognizes two MBNL1 isoforms generated by alternative splicing (∼41 and 42 kDa). GAPDH (∼36 kDa) was used as a loading control. (B) Immunofluorescence using mAb 3A4 to confirm depletion of endogenous protein after independent transfection of each MBNL1 siRNA construct. Scale bar, 10 μm. (C) siRNA-mediated depletion of MBNL1 with two independent constructs reproduces the DM splicing patterns for cTNT and IR minigenes. RT–PCR results are from at least three transfections. GFP siRNA had no effect on splicing of any of the tested minigenes. MBNL1 siRNA had minimal effects on splicing of a rat clathrin light-chain minigene.
Figure 3
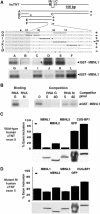
MBNL1 binds upstream of exon 5 in human cTNT at a site distinct from the CUG-BP1-binding site. (A) Binding of recombinant GST–MBNL1 to uniformly 32P-labeled RNA was assayed by UV crosslinking. Scanning mutagenesis was performed by replacing 6 nt blocks with AUAAUA and identified two binding sites 18 and 36 nt upstream of the alternative exon. The MBNL1-binding sites (M) and the CUG-BP1-binding site (C) are located on opposite sides of exon 5. (+) and (−) indicate binding; (•) indicates a putative branch point adenosine. (B) Four nucleotide substitutions significantly reduce binding of recombinant MBNL1 detected by UV crosslinking. Competition of GST–MBNL1 binding to 32P-labeled RNA G by the indicated picomoles of nonlabeled RNAs G or M shown in A). (C, D) MBNL1-binding site mutations reduce responsiveness to MBNL1, MBNL2 and MBNL3 coexpression but not CUG-BP1 in COSM6 cells. Human cTNT minigenes containing the natural sequence (C) or the four nucleotide substitutions (mutation M in A) in the MBNL1-binding site (D) were coexpressed with GFP or the indicated GFP fusion proteins. Exon inclusion was assayed by RT–PCR.
Figure 4
MBNL1 binds to _cis_-elements in chicken cTNT intron 5 required for muscle-specific splicing. (A) The chicken cTNT MSE1–4 RNA contains an alternative exon flanked by four MSEs. GST–MBNL1 bound weakly to MSE1 and strongly to MSE4 in UV-crosslinking assays. (B) Competition of GST–MBNL1 binding to labeled chicken cTNT MSE1–4 RNA by nonlabeled MSE RNAs. Picomoles of competitor RNA are indicated. (C) Scanning mutagenesis identified two MBNL1-binding sites within MSE4. (D) Alignment of the four MBNL1-binding motifs in human and chicken cTNT reveals a common motif.
Figure 5
Regulation of human cTNT by MBNL1 is independent of CELF regulation. The (A) wild-type cTNT minigene or a (B) mutant cTNT minigene with point mutations that prevent CUG-BP1 binding and regulation were cotransfected with the indicated siRNA constructs, a plasmid expressing a DMPK minigene with 960 CUG repeats (Philips et al, 1998) or a GFP–MBNL1 expression plasmid in HeLa cells.
Figure 6
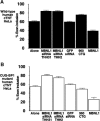
Deletion of the human IR CUG-BP1-binding site does not affect regulation by MBNL1. (A) All the three MBNL proteins promote exon 11 inclusion of a mutant human IR minigene lacking the CUG-BP1-binding site in HEK293 cells. (B) RNAi depletion of MBNL1 in HeLa cells using the indicated siRNA constructs promotes exon 11 skipping in a human IR minigene lacking the CUG-BP1-binding site.
Similar articles
- Alternative splicing of the neurofibromatosis type 1 pre-mRNA is regulated by the muscleblind-like proteins and the CUG-BP and ELAV-like factors.
Fleming VA, Geng C, Ladd AN, Lou H. Fleming VA, et al. BMC Mol Biol. 2012 Dec 10;13:35. doi: 10.1186/1471-2199-13-35. BMC Mol Biol. 2012. PMID: 23227900 Free PMC article. - The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing.
Ladd AN, Charlet N, Cooper TA. Ladd AN, et al. Mol Cell Biol. 2001 Feb;21(4):1285-96. doi: 10.1128/MCB.21.4.1285-1296.2001. Mol Cell Biol. 2001. PMID: 11158314 Free PMC article. - MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T.
Warf MB, Berglund JA. Warf MB, et al. RNA. 2007 Dec;13(12):2238-51. doi: 10.1261/rna.610607. Epub 2007 Oct 17. RNA. 2007. PMID: 17942744 Free PMC article. - The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing.
Pascual M, Vicente M, Monferrer L, Artero R. Pascual M, et al. Differentiation. 2006 Mar;74(2-3):65-80. doi: 10.1111/j.1432-0436.2006.00060.x. Differentiation. 2006. PMID: 16533306 Review. - MBNL proteins and their target RNAs, interaction and splicing regulation.
Konieczny P, Stepniak-Konieczna E, Sobczak K. Konieczny P, et al. Nucleic Acids Res. 2014;42(17):10873-87. doi: 10.1093/nar/gku767. Epub 2014 Sep 2. Nucleic Acids Res. 2014. PMID: 25183524 Free PMC article. Review.
Cited by
- CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease.
Ladd AN. Ladd AN. Mol Cell Neurosci. 2013 Sep;56:456-64. doi: 10.1016/j.mcn.2012.12.003. Epub 2012 Dec 14. Mol Cell Neurosci. 2013. PMID: 23247071 Free PMC article. Review. - Molecular mechanisms in DM1 - a focus on foci.
Pettersson OJ, Aagaard L, Jensen TG, Damgaard CK. Pettersson OJ, et al. Nucleic Acids Res. 2015 Feb 27;43(4):2433-41. doi: 10.1093/nar/gkv029. Epub 2015 Jan 20. Nucleic Acids Res. 2015. PMID: 25605794 Free PMC article. Review. - A biophysical model for identifying splicing regulatory elements and their interactions.
Wen J, Chen Z, Cai X. Wen J, et al. PLoS One. 2013;8(1):e54885. doi: 10.1371/journal.pone.0054885. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23382993 Free PMC article. - Insulin Receptor Isoforms in Cancer.
Vella V, Milluzzo A, Scalisi NM, Vigneri P, Sciacca L. Vella V, et al. Int J Mol Sci. 2018 Nov 16;19(11):3615. doi: 10.3390/ijms19113615. Int J Mol Sci. 2018. PMID: 30453495 Free PMC article. Review. - Autophagy regulation by RNA alternative splicing and implications in human diseases.
González-Rodríguez P, Klionsky DJ, Joseph B. González-Rodríguez P, et al. Nat Commun. 2022 May 18;13(1):2735. doi: 10.1038/s41467-022-30433-1. Nat Commun. 2022. PMID: 35585060 Free PMC article. Review.
References
- Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, Baylies MK (1998) The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev Biol 195: 131–143 - PubMed
- Begemann G, Paricio N, Artero R, Kiss I, Perez-Alonso M, Mlodzik M (1997) Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development 124: 4321–4331 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AR46799/AR/NIAMS NIH HHS/United States
- AR45653/AR/NIAMS NIH HHS/United States
- P50 NS048843/NS/NINDS NIH HHS/United States
- R01 AR046799/AR/NIAMS NIH HHS/United States
- R01 AR045653/AR/NIAMS NIH HHS/United States
- HL45565/HL/NHLBI NIH HHS/United States
- NS48843/NS/NINDS NIH HHS/United States
- R01 HL045565/HL/NHLBI NIH HHS/United States
- U54 NS048843/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials