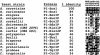Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species - PubMed (original) (raw)
. 2004 Jul 27;32(13):3947-56.
doi: 10.1093/nar/gkh734. Print 2004.
Affiliations
- PMID: 15280510
- PMCID: PMC506816
- DOI: 10.1093/nar/gkh734
Evolution of divergent DNA recognition specificities in VDE homing endonucleases from two yeast species
Karen L Posey et al. Nucleic Acids Res. 2004.
Abstract
Homing endonuclease genes (HEGs) are mobile DNA elements that are thought to confer no benefit to their host. They encode site-specific DNA endonucleases that perpetuate the element within a species population by homing and disseminate it between species by horizontal transfer. Several yeast species contain the VMA1 HEG that encodes the intein-associated VMA1-derived endonuclease (VDE). The evolutionary state of VDEs from 12 species was assessed by assaying their endonuclease activities. Only two enzymes are active, PI-ZbaI from Zygosaccharomyces bailii and PI-ScaI from Saccharomyces cariocanus. PI-ZbaI cleaves the Z.bailii recognition sequence significantly faster than the Saccharomyces cerevisiae site, which differs at six nucleotide positions. A mutational analysis indicates that PI-ZbaI cleaves the S.cerevisiae substrate poorly due to the absence of a contact that is analogous to one made in PI-SceI between Gln-55 and nucleotides +9/+10. PI-ZbaI cleaves the Z.bailii substrate primarily due to a single base-pair substitution (A/T+5 --> T/A+5). Structural modeling of the PI-ZbaI/DNA complex suggests that Arg-331, which is absent in PI-SceI, contacts T/A+5, and the reduced activity observed in a PI-ZbaI R331A mutant provides evidence for this interaction. These data illustrate that homing endonucleases evolve altered specificity as they adapt to recognize alternative target sites.
Figures
Figure 1
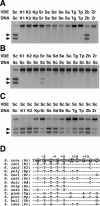
Assay of endonuclease cleavage activity of DNA substrates containing single recognition sites by VDE proteins. The reactions were performed by combining purified protein (100 nM) with duplex DNA substrates (7 nM) for various lengths of time at 37°C. (A) Cleavage of DNA substrates containing the predicted VDE target sites from the different yeast species after 6 h by their respective VDE proteins (species abbreviations are as follows: Sc, S.cerevisiae; Sr, S.cariocanus; Se, S.exiguus; Ss, S.castellii; Sd, S.dairensis; Su, S.unisporus; Kp, K.polysporus; Zb, Z.bailii; Zr, Z.rouxii; Tp, T.pretoriensis; Tg, T.globosa; K1, K.lactis strain CBS 2896; and K2, K.lactis strain CBS 683). Arrows indicate the position of the two DNA cleavage products. (B) Cleavage of a DNA substrate containing the S.cerevisiae recognition site after 6 h of incubation with different VDE proteins. (C) Cleavage of DNA substrates containing the different yeast VDE recognition sites after 1 h by PI-SceI. (D) A comparison of the DNA sequences of the different predicted target sites of each yeast species. The top strand of the S.cerevisiae recognition site is shown on the top line with the base-pair positions relative to the center of the 4 bp cleavage site overhang, as indicated above. Below are shown the aligned bases that differ from the S.cerevisiae sequence. The highlighted bases are those that differ in one or more of the yeast species.
Figure 2
Sequence alignment of active site residues in PI-SceI with those of other yeast VDEs. The percent identity between the different VDE sequences and PI-SceI is given. Active site residues that are identical at the aligned positions are highlighted. The positions of the PI-SceI residues in the primary sequence are indicated above each position. Asp-218 and Asp-326 are two conserved amino acids that coordinate the essential divalent metal ions and are critical for activity. The exact roles of Asp-229, Lys-301, Thr-341 and Lys-403 in catalysis have not been defined, but their substitution with other amino acids decreases the PI-SceI catalytic activity (27).
Figure 3
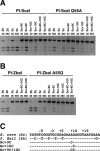
Assay of endonuclease cleavage activity of wild-type PI-SceI, PI-SceI Q55A, wild-type PI-ZbaI and PI-ZbaI A55Q proteins on DNA substrates containing a single recognition site. In each reaction, the protein (100 nM) was incubated with DNA substrate (7 nM) in Mg2+ cleavage buffer at 37°C. (A) DNA cleavage of wild-type and mutant substrates (indicated above each lane) by wild-type PI-SceI and PI-SceI Q55A proteins. DNA substrates containing the S.cerevisiae and Z.bailii recognition sites are denoted as Sc and Zb, respectively, followed by any introduced substitutions. (B) DNA cleavage assay of wild-type PI-ZbaI and PI-ZbaI A55Q proteins using wild-type and mutant DNA substrates. (C) The sequences of the different DNA substrates.
Figure 4
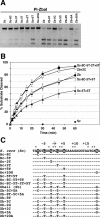
Identification of the base pairs in the Z.bailii substrate that contribute to PI-ZbaI activity. (A) Plus-ones/minus-ones analysis of the PI-ZbaI activity. DNA substrates (7 nM) containing individually substituted Z.bailii base pairs within the S.cerevisiae recognition site (plus-ones) or containing individually substituted S.cerevisiae base pairs within the Z.bailii recognition site (minus-ones) were incubated at 37°C for 6 h with PI-ZbaI (100 nM). (B) Time course of DNA cleavage of wild-type and mutant substrates by PI-ZbaI. The percentages of product formed by digestion of the S.cerevisiae (filled diamonds), Sc-5T+5T (filled triangles), Z.bailii (filled circles), Sc-8C-5T+5T (open circles), Sc-8C-5T-2T+5T (filled squares) and Zb+2C (open squares) substrates are plotted as a function of time. (C) The DNA sequences of the different DNA substrates used in this study. The S.cerevisiae target base pairs indicated by an asterisk are those that are important for substrate cleavage by PI-SceI. The base pairs in the VDE target sequences that are investigated in this experiment are highlighted.
Figure 5
Cleavage of DNA substrates containing the wild-type or Zb+5A recognition site by wild-type PI-ZbaI, PI-ZbaI R331A and PI-ZbaI H329A proteins. Protein (100 nM) was incubated with substrate (7 nM) at 37°C for either 1 h in buffer containing Mn2+ or 6 h in buffer containing Mg2+. The DNA sequences of these substrates are indicated in Figure 4C.
Similar articles
- Occurrence, horizontal transfer and degeneration of VDE intein family in Saccharomycete yeasts.
Okuda Y, Sasaki D, Nogami S, Kaneko Y, Ohya Y, Anraku Y. Okuda Y, et al. Yeast. 2003 May;20(7):563-73. doi: 10.1002/yea.984. Yeast. 2003. PMID: 12734795 - Evolution of I-SceI homing endonucleases with increased DNA recognition site specificity.
Joshi R, Ho KK, Tenney K, Chen JH, Golden BL, Gimble FS. Joshi R, et al. J Mol Biol. 2011 Jan 7;405(1):185-200. doi: 10.1016/j.jmb.2010.10.029. Epub 2010 Oct 26. J Mol Biol. 2011. PMID: 21029741 Free PMC article. - Novel site-specific DNA endonucleases.
Aggarwal AK, Wah DA. Aggarwal AK, et al. Curr Opin Struct Biol. 1998 Feb;8(1):19-25. doi: 10.1016/s0959-440x(98)80005-5. Curr Opin Struct Biol. 1998. PMID: 9519292 Review. - Invasion of a multitude of genetic niches by mobile endonuclease genes.
Gimble FS. Gimble FS. FEMS Microbiol Lett. 2000 Apr 15;185(2):99-107. doi: 10.1111/j.1574-6968.2000.tb09046.x. FEMS Microbiol Lett. 2000. PMID: 10754232 Review.
Cited by
- Investigation of the mechanism of meiotic DNA cleavage by VMA1-derived endonuclease uncovers a meiotic alteration in chromatin structure around the target site.
Fukuda T, Ohta K, Ohya Y. Fukuda T, et al. Eukaryot Cell. 2006 Jun;5(6):981-90. doi: 10.1128/EC.00052-06. Eukaryot Cell. 2006. PMID: 16757746 Free PMC article. - Nuclease genes occupy boundaries of genetic exchange between bacteriophages.
Barth ZK, Dunham DT, Seed KD. Barth ZK, et al. NAR Genom Bioinform. 2023 Aug 24;5(3):lqad076. doi: 10.1093/nargab/lqad076. eCollection 2023 Sep. NAR Genom Bioinform. 2023. PMID: 37636022 Free PMC article. - Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation.
Takeuchi R, Certo M, Caprara MG, Scharenberg AM, Stoddard BL. Takeuchi R, et al. Nucleic Acids Res. 2009 Feb;37(3):877-90. doi: 10.1093/nar/gkn1007. Epub 2008 Dec 22. Nucleic Acids Res. 2009. PMID: 19103658 Free PMC article. - Evolutionary maintenance of selfish homing endonuclease genes in the absence of horizontal transfer.
Yahara K, Fukuyo M, Sasaki A, Kobayashi I. Yahara K, et al. Proc Natl Acad Sci U S A. 2009 Nov 3;106(44):18861-6. doi: 10.1073/pnas.0908404106. Epub 2009 Oct 16. Proc Natl Acad Sci U S A. 2009. PMID: 19837694 Free PMC article. - Evolution and Application of Inteins in Candida species: A Review.
Fernandes JA, Prandini TH, Castro MD, Arantes TD, Giacobino J, Bagagli E, Theodoro RC. Fernandes JA, et al. Front Microbiol. 2016 Oct 10;7:1585. doi: 10.3389/fmicb.2016.01585. eCollection 2016. Front Microbiol. 2016. PMID: 27777569 Free PMC article. Review.
References
- Gimble F.S. (2000) Invasion of a multitude of genetic niches by homing endonuclease genes. FEMS Microbiol. Lett., 185, 99–107. - PubMed
- Garrett R.A., Dalgaard,J., Larsen,N., Kjems,J. and Mankin,A.S. (1991) Archaeal rRNA operons. Trends Biochem. Sci., 16, 22–26. - PubMed
- Dalgaard J.Z., Moser,M.J., Hughey,R. and Mian,I.S. (1997) Statistical modeling, phylogenetic analysis and structure prediction of a protein splicing domain common to inteins and hedgehog proteins. J. Comput. Biol., 4, 193–214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous