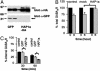Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type A receptor membrane trafficking - PubMed (original) (raw)
Huntingtin-associated protein 1 regulates inhibitory synaptic transmission by modulating gamma-aminobutyric acid type A receptor membrane trafficking
Josef T Kittler et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Gamma-aminobutyric acid type A receptors (GABA(A)Rs) are the major sites of fast synaptic inhibition in the brain. An essential determinant for the efficacy of synaptic inhibition is the regulation of GABA(A)R cell surface stability. Here, we have examined the regulation of GABA(A)R endocytic sorting, a critical regulator of cell surface receptor number. In neurons, rapid constitutive endocytosis of GABA(A)Rs was evident. Internalized receptors were then either rapidly recycled back to the cell surface, or on a slower time scale, targeted for lysosomal degradation. This sorting decision was regulated by a direct interaction of GABA(A)Rs with Huntingtin-associated protein 1 (HAP1). HAP1 modulated synaptic GABA(A)R number by inhibiting receptor degradation and facilitating receptor recycling. Together these observations have identified a role for HAP1 in regulating GABA(A)R sorting, suggesting an important role for this protein in the construction and maintenance of inhibitory synapses.
Figures
Fig. 1.
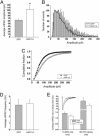
Endocytosis and recycling of GABAARs in cortical neurons. (A) Analysis of receptor endocytosis in cortical neurons. Cortical neurons were surface biotnylated and incubated for 30 min at 4°C to inhibit membrane traffic (lanes 1 and 2) or 37°C to allow endocytosis (lane 3). Cells were treated with glutathione to cleave remaining cell surface biotin (lanes 2 and 3). Cells were then lysed, and biotinylated proteins were purified on beads immobilized to streptavidin, separated by SDS/PAGE followed by quantitative immunoblotting with anti-β3 antibody. Lane 1 represents total cell surface receptor levels, lane 2 represents background internalization at 4°C (control), and lane 3 represents protected biotinylated receptors internalized at 37°C for 30 min followed by cleavage with glutathione. Molecular markers in kDa are shown. (B) Time course of GABAAR endocytosis. Receptors internalized after various times at 37°C were compared to total surface receptor levels to quantify percent internalization (n = 4-7). (Inset) A representative Western blot. (C) Recycling of internalized GABAARs. (Inset) Representative Western blot of recycling time course. Cortical neurons were labeled with biotin and receptors were allowed to internalize for 30 min, before cleavage with glutathione (as in A and B). Neurons were then incubated at 37°C for further time periods (lanes 2-6, representing 0, 5, 15, 30, and 60 min, respectively) with glutathione in the external culture medium to cleave any internalized biotinylated receptors recycling to the cell surface. Internal β3-subunit levels remaining, as measured with anti-β3 antibody by Western blotting, were then compared to those at the start of the second incubation period with glutathione (lane 2, time = 0 min), designated as 100%. No loss of biotinylated β3 subunit (i.e., recycling) was observed if intracellular transport was blocked at 4°C (lane 7), or when neurons were incubated at 37°C in the absence of glutathione in the external media (lane 1). (D) Analysis of GABAAR degradation. Quantitation of GABAAR degradation (loss of biotinylated receptor represents degradation) expressed as the percent of total surface biotinylated GABAAR remaining at different incubation times (n = 3-5) in the presence (gray bar) or absence (black bar) of leupeptin (200 μg/ml) used to inhibit the proteolytic activity of lysosomes. * indicates significantly different (P < 0.05).
Fig. 2.
Specific interaction of GABAAR β subunits with HAP1. (A) HAP1 interacts specifically with GABAAR β1 subunit in the Y2H system. Interaction between the HAP1 Y2H clone and various GABAAR subunit intracellular domains was assayed by Y2H and β-galactosidase assay. R1, GABABR1 subunit; +, positive interaction; -, no interaction detected; v, empty vector. (B) The direct binding of HAP1a with the intracellular domains of GABAAR β1 subunit was examined. The GABABR1 subunit C-terminal tail expressed as GST fusion protein (lane 2), the intracellular domain of the GABAAR β1 subunit (lane 3), or GST alone (lane 4) immobilized on glutathione agarose were exposed to 35S-methionine-radiolabeled HAP1a. Bound material was separated by SDS/PAGE and visualized by autoradiography. Lane 1 represents 25% of 35S-methionine-radiolabeled HAP1a used in pull-down assay. (C) Further analysis of HAP1 specificity was carried out with GST fusion protein pull-down assays from COS cell lysates expressing HAP1b (lane 1) with the intracellular domains of the GABAAR: α1 (lane 2), α3 (lane 3), α6 (lane 4), β1 (lane 5), β2 (lane 6), β3 (lane 7), γ2S (lane 8), δ (lane 9), and GABAC receptor ρ1 (lane 10) expressed as GST fusion proteins or GST alone (lane 11). HAP1b binding was then measured by immunoblotting. Lane 1 represents 25% of the input used for each experiment. (D) GST constructs encoding the intracellular domains of the GABAAR α1 (lane 2), β1 (lane 3), β2 (lane 4), β3 lane (lane 5), γ2S (lane 6), or GST alone (lane 7) were immobilized on glutathione agarose and incubated with solubilized brain extracts. Bound material was resolved by SDS/PAGE and analyzed by Western blotting with antibodies to HAP1a and HAP1b. Lane 1 represents 25% of the material used in each experiment. (E) Solubilized brain homogenates were subjected to immunoprecipitation with anti-GABAAR antibodies to the β1 and β3 subunits (lane 2) or control IgG (lane 3). Immunoprecipitates were immunoblotted with mouse HAP1a/b antibody. Lane 1 represents 10% of the input used in each experiment. (F) Solubilized brain extracts were immunoprecipitated with antibodies against HAP1 (lane 2) or control IgG (lane 3). Precipitated material was then immunoblotted with antibodies against HAP1 or the GABAAR β2 and β3 subunits as indicated. Lane 1 represents 10% of the input. (G) Colocalization of HAP1 with GABAARs. Hippocampal neurons (21 DIV) were immunostained for the receptor γ2 subunit (1) and HAP1 (2). A merged image is shown in 3. Arrows identify colocalized clusters of HAP1 and GABAARs in the soma and dendritic shafts of hippocampal pyramidal neurons. The data are single 0.5-μm optical sections. (Scale bar: 20 μm.)
Fig. 3.
Recycling and degradation of GABAARs in cortical neurons is modulated by HAP1. (A) Representative Western blot of lysates from cortical neurons nucleofected with GFP or HA-HAP1a showing efficient expression of exogenous cDNAs (arrow) in neurons by nucleofection. Using antibody against the HA epitope a nonspecific band was also detected. Molecular markers in kDa are shown. (B) Biotinylation assays of receptor degradation were performed (as in Fig. 1_D_) in cortical neurons either mock-nucleofected or nucleofected with HA-HAP1a. Quantitation of GABAAR degradation is expressed as the percent of the total surface biotinylated GABAAR pool at time 0 remaining after 6 h. Expression of HA-HAP1a in neurons produced a significant decrease in GABAAR degradation compared to mock-nucleofected neurons as indicated by * (P < 0.05, n = 7). Control data represent receptor degradation in nonnucleofected neurons (from Fig. 1_D_). (C) Recycling of internalized GABAARs is modulated by overexpressing HAP1 in cortical neurons. Biotinylation assays of GABAAR recycling (for 30 or 60 min after a 30-min endocytosis period) were performed (as in Fig. 1_C_) on cortical neurons nucleofected with either GFP or HA-HAP1a. The loss of biotinylated GABAARs after a second biotin cleavage provides a measure of receptor recycling. Data for GABAAR recycling of nonnucleofected neurons at 30- and 60-min time points (from Fig. 1_C_) are presented as the control column. Expression of HA-HAP1a in neurons produced a significant increase in GABAAR recycling compared to GFP-nucleofected neurons as indicated by * (P < 0.05, n = 7).
Fig. 4.
mIPSC amplitudes are increased upon expression of HAP-1a protein. (A) Bar graph of the mean mIPSC amplitudes representing GFP (hatched bar) or GFP+HA-HAP1a (open bar) cDNA-transfected hippocampal neurons. Data (mean ± SE) are pooled from seven cells for each condition on 14-DIV hippocampal neuronal cultures. * indicates significance (P < 0.05). (B) Peak amplitude binned data plots for mIPSCs recorded from neurons expressing GFP (solid line) or GFP+HAP-1a (open bars). Note that the distribution for the mIPSCs recorded from HA-HAP1a neurons are shifted to higher amplitudes relative to neurons expressing GFP alone. (C) Cumulative probability data for mIPSC amplitudes demonstrating the significant amplitude shift between GFP-transfected (filled symbols) and GFP+HAP-1a-transfected (open symbols) neurons (P < 0.001). (D) Bar graph of the mean mIPSC frequencies recorded from neurons expressing GFP (n = 8) or GFP+HA-HAP-1a (n = 12) from five independent cultures. The frequency measurements were made over 14 min of consecutive recording from each transfected cell. (E) The presence of HA-HAP-1a protein (open bar) did not affect the kinetic profile of the mIPSCs as both rise (10-90%) and decay (90-37%) times (msec) were not significantly different from GFP (hatched bar) controls [P = 0.12 (rise, n = 9) and 0.18 (decay, n = 13)]. (Inset) Scaled average mIPSC profiles (from 50 mIPSCs) for GFP-transfected (left trace) and HA-HAP-1a-transfected (right trace) cells.
Fig. 5.
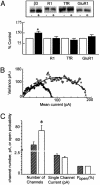
HAP1 controls GABAAR cell surface and synaptic number. (A) Control cortical neurons expressing GFP (-) or neurons expressing HA-HAP1a (+) were biotinylated. Cells were then lysed, and cell surface biotinylated receptor populations were purified on beads immobilized to streptavidin, separated by SDS/PAGE, followed by quantitative immunoblotting with antibodies against the GABAAR β3 subunit (β3), the GABABR1 subunit (R1), the TfR, and the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor GluR1 subunit (GluR1) (Upper). Cell surface receptor levels for control (open bar) and HA-HAP1a-expressing neurons (filled bar) were then compared for each of these proteins. * indicates significantly different from control (P < 0.05, n = 5-9). The number of GABAARs in neurons expressing HA-HAP1a was enhanced to 145 ± 11% of control (P < 0.05, n = 9). (B) Nonstationary noise analysis of mIPSCs. Current-variance relationships for two example neurons (14 DIV) transfected either with GFP (•) or HAP1 cDNAs (○). Solid lines depict the theoretical fits generated by the equation in Materials and Methods. (C) Bar graph showing the relative numbers of synaptic GABAARs that are activated at the peak of an average mIPSC event in either control GFP-transfected (hatched bar) or HAP1-transfected (open bar) hippocampal neurons (14 DIV, n = 4 individual experiments, P < 0.05). Also shown are the relative sizes of single channel currents (pA) and the probability of channel opening (_P_open).
Similar articles
- Calcium-modulating cyclophilin ligand regulates membrane trafficking of postsynaptic GABA(A) receptors.
Yuan X, Yao J, Norris D, Tran DD, Bram RJ, Chen G, Luscher B. Yuan X, et al. Mol Cell Neurosci. 2008 Jun;38(2):277-89. doi: 10.1016/j.mcn.2008.03.002. Epub 2008 Mar 13. Mol Cell Neurosci. 2008. PMID: 18424167 Free PMC article. - Delivery of GABAARs to synapses is mediated by HAP1-KIF5 and disrupted by mutant huntingtin.
Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, Kittler JT. Twelvetrees AE, et al. Neuron. 2010 Jan 14;65(1):53-65. doi: 10.1016/j.neuron.2009.12.007. Neuron. 2010. PMID: 20152113 Free PMC article. - Constitutive endocytosis of GABAA receptors by an association with the adaptin AP2 complex modulates inhibitory synaptic currents in hippocampal neurons.
Kittler JT, Delmas P, Jovanovic JN, Brown DA, Smart TG, Moss SJ. Kittler JT, et al. J Neurosci. 2000 Nov 1;20(21):7972-7. doi: 10.1523/JNEUROSCI.20-21-07972.2000. J Neurosci. 2000. PMID: 11050117 Free PMC article. - The role of GABAAR phosphorylation in the construction of inhibitory synapses and the efficacy of neuronal inhibition.
Vithlani M, Moss SJ. Vithlani M, et al. Biochem Soc Trans. 2009 Dec;37(Pt 6):1355-8. doi: 10.1042/BST0371355. Biochem Soc Trans. 2009. PMID: 19909275 Free PMC article. Review. - GABA A,slow: causes and consequences.
Capogna M, Pearce RA. Capogna M, et al. Trends Neurosci. 2011 Feb;34(2):101-12. doi: 10.1016/j.tins.2010.10.005. Epub 2010 Dec 9. Trends Neurosci. 2011. PMID: 21145601 Review.
Cited by
- Rapid suppression of inhibitory synaptic transmission by retinoic acid.
Sarti F, Zhang Z, Schroeder J, Chen L. Sarti F, et al. J Neurosci. 2013 Jul 10;33(28):11440-50. doi: 10.1523/JNEUROSCI.1710-13.2013. J Neurosci. 2013. PMID: 23843516 Free PMC article. - Gephyrin: a key regulatory protein of inhibitory synapses and beyond.
Groeneweg FL, Trattnig C, Kuhse J, Nawrotzki RA, Kirsch J. Groeneweg FL, et al. Histochem Cell Biol. 2018 Nov;150(5):489-508. doi: 10.1007/s00418-018-1725-2. Epub 2018 Sep 27. Histochem Cell Biol. 2018. PMID: 30264265 Review. - Ubiquitin-dependent lysosomal targeting of GABA(A) receptors regulates neuronal inhibition.
Arancibia-Cárcamo IL, Yuen EY, Muir J, Lumb MJ, Michels G, Saliba RS, Smart TG, Yan Z, Kittler JT, Moss SJ. Arancibia-Cárcamo IL, et al. Proc Natl Acad Sci U S A. 2009 Oct 13;106(41):17552-7. doi: 10.1073/pnas.0905502106. Epub 2009 Oct 6. Proc Natl Acad Sci U S A. 2009. PMID: 19815531 Free PMC article. - Agonist-dependent endocytosis of γ-aminobutyric acid type A (GABAA) receptors revealed by a γ2(R43Q) epilepsy mutation.
Chaumont S, André C, Perrais D, Boué-Grabot E, Taly A, Garret M. Chaumont S, et al. J Biol Chem. 2013 Sep 27;288(39):28254-65. doi: 10.1074/jbc.M113.470807. Epub 2013 Aug 9. J Biol Chem. 2013. PMID: 23935098 Free PMC article. - Distinct mechanisms drive sequential internalization and degradation of GABAARs during global ischemia and reperfusion injury.
Garcia JD, Wolfe SE, Stewart AR, Tiemeier E, Gookin SE, Guerrero MB, Quillinan N, Smith KR. Garcia JD, et al. iScience. 2023 Sep 27;26(10):108061. doi: 10.1016/j.isci.2023.108061. eCollection 2023 Oct 20. iScience. 2023. PMID: 37860758 Free PMC article.
References
- Moss, S. J. & Smart, T. G. (2001) Nat. Rev. Neurosci. 2, 240-250. - PubMed
- Sieghart, W. & Sperk, G. (2002) Curr. Top. Med. Chem. 2, 795-816. - PubMed
- Kittler, J. T. & Moss, S. J. (2001) Traffic 2, 437-448. - PubMed
- Nusser, Z., Hajos, N., Somogyi, P. & Mody, I. (1998) Nature 395, 172-177. - PubMed
- Nusser, Z., Cull-Candy, S. & Farrant, M. (1997) Neuron 19, 697-709. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous