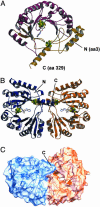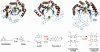Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans - PubMed (original) (raw)
Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans
Petra Hänzelmann et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The MoaA and MoaC proteins catalyze the first step during molybdenum cofactor biosynthesis, the conversion of a guanosine derivative to precursor Z. MoaA belongs to the S-adenosylmethionine (SAM)-dependent radical enzyme superfamily, members of which catalyze the formation of protein and/or substrate radicals by reductive cleavage of SAM by a [4Fe-4S] cluster. A defined in vitro system is described, which generates precursor Z and led to the identification of 5'-GTP as the substrate. The structures of MoaA in the apo-state (2.8 angstroms) and in complex with SAM (2.2 angstroms) provide valuable insights into its mechanism and help to define the defects caused by mutations in the human ortholog of MoaA that lead to molybdenum cofactor deficiency, a usually fatal disease accompanied by severe neurological symptoms. The central core of each subunit of the MoaA dimer is an incomplete triosephosphate isomerase barrel formed by the N-terminal part of the protein, which contains the [4Fe-4S] cluster typical for SAM-dependent radical enzymes. SAM is the fourth ligand to the cluster and binds to its unique Fe as an N/O chelate. The lateral opening of the incomplete triosephosphate isomerase barrel is covered by the C-terminal part of the protein containing an additional [4Fe-4S] cluster, which is unique to MoaA proteins. Both FeS clusters are separated by approximately 17 angstroms, with a large active site pocket between. The noncysteinyl-ligated unique Fe site of the C-terminal [4Fe-4S] cluster is proposed to be involved in the binding and activation of 5'-GTP.
Copyright 2004 The National Academy of Sciencs of the USA
Figures
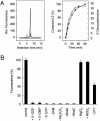
Fig. 1.
In vitro synthesis of precursor Z. (A Left) Synthesis of precursor Z determined by measurement of its stable oxidized derivative, compound Z, by reversed-phase HPLC. MoaA (50 μM) and MoaC (50 μM) were incubated under anaerobic conditions in the presence of 2 mM DTT with a 10-fold molar excess of SAM, 5′-GTP, MgCl2, and Na2S2O4 for 45 min (standard assay). (Top to bottom) Reconstituted MoaA; anaerobically purified MoaA; without MoaC. (Right) Time dependence of precursor Z formation (•) and reductive cleavage of SAM in the presence of 5′-GTP generating 5′-dA (○). (B) Requirements for precursor Z synthesis. The maximal amount of compound Z formed in a standard assay (control, see A) was set to 100%. 5′-GTP was absent from the experiment with 5′-GDP and 5′-GMP.
Fig. 2.
Overall structure of MoaA. (A) Ribbon diagram of the monomer. The N-terminal TIM barrel structure is shown in maroon, and the C-terminal part is shown in gold. (B) Ribbon diagram of the dimer. The two monomers are colored in blue and orange. (C) Molecular surface of the dimer colored as in B viewed along the hydrophilic channel in one of the monomers. FeS clusters are shown in
cpk
(Fe, green; S, yellow), and SAM is shown in stick representation.
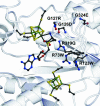
Fig. 3.
The active site of MoaA. (A) Stereoview of the FeS clusters and SAM. SAM is shown in a _F_o–_F_c omit map contoured at 3 σ. Hydrogen bonds are indicated as dashed lines. (B) Closer view of the active-site pocket created by predominantly positively charged amino acids. Conserved amino acids are labeled in red. Secondary structural elements and cofactors are rendered transparent. (C) Surface representation viewed from both sides into the active site channel. Conserved residues are mapped onto the molecular surface. (D) σA weighted 2_F_o–_F_c map of the C-terminal FeS cluster with a DTT molecule as the fourth ligand contoured at 1 σ. Surrounding amino acids are shown as a Cα trace colored in orange. Amino acids, FeS clusters, and SAM are shown in ball and stick.
Fig. 4.
Location of missense mutations detected in Moco-deficient patients. Secondary structure elements and cofactors are rendered transparent. Human mutations, FeS clusters, and SAM are shown as ball and stick.
Fig. 5.
Structural comparison of SAM-dependent radical enzymes and reactions catalyzed. α-Helices are colored in orange, β-sheets are colored in blue, and turns are colored in gray. Nonstructurally conserved secondary structural elements are rendered transparent. FeS clusters and substrates are shown in ball and stick.
Similar articles
- SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes.
Grell TA, Goldman PJ, Drennan CL. Grell TA, et al. J Biol Chem. 2015 Feb 13;290(7):3964-71. doi: 10.1074/jbc.R114.581249. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477505 Free PMC article. Review. - Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism.
Hänzelmann P, Schindelin H. Hänzelmann P, et al. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6829-34. doi: 10.1073/pnas.0510711103. Epub 2006 Apr 21. Proc Natl Acad Sci U S A. 2006. PMID: 16632608 Free PMC article. - Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8.
Kanaujia SP, Jeyakanthan J, Nakagawa N, Balasubramaniam S, Shinkai A, Kuramitsu S, Yokoyama S, Sekar K. Kanaujia SP, et al. Acta Crystallogr D Biol Crystallogr. 2010 Jul;66(Pt 7):821-33. doi: 10.1107/S0907444910019074. Epub 2010 Jun 19. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20606263 - Characterization of MOCS1A, an oxygen-sensitive iron-sulfur protein involved in human molybdenum cofactor biosynthesis.
Hänzelmann P, Hernández HL, Menzel C, García-Serres R, Huynh BH, Johnson MK, Mendel RR, Schindelin H. Hänzelmann P, et al. J Biol Chem. 2004 Aug 13;279(33):34721-32. doi: 10.1074/jbc.M313398200. Epub 2004 Jun 4. J Biol Chem. 2004. PMID: 15180982 - Radical Breakthroughs in Natural Product and Cofactor Biosynthesis.
Yokoyama K. Yokoyama K. Biochemistry. 2018 Jan 30;57(4):390-402. doi: 10.1021/acs.biochem.7b00878. Epub 2017 Nov 9. Biochemistry. 2018. PMID: 29072833 Free PMC article. Review.
Cited by
- Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system.
King PW, Posewitz MC, Ghirardi ML, Seibert M. King PW, et al. J Bacteriol. 2006 Mar;188(6):2163-72. doi: 10.1128/JB.188.6.2163-2172.2006. J Bacteriol. 2006. PMID: 16513746 Free PMC article. - Structure and stability of the molybdenum cofactor intermediate cyclic pyranopterin monophosphate.
Santamaria-Araujo JA, Wray V, Schwarz G. Santamaria-Araujo JA, et al. J Biol Inorg Chem. 2012 Jan;17(1):113-22. doi: 10.1007/s00775-011-0835-2. Epub 2011 Aug 30. J Biol Inorg Chem. 2012. PMID: 21877100 - An unusual genetic variant in the MOCS1 gene leads to complete missplicing of an alternatively spliced exon in a patient with molybdenum cofactor deficiency.
Arenas M, Fairbanks LD, Vijayakumar K, Carr L, Escuredo E, Marinaki AM. Arenas M, et al. J Inherit Metab Dis. 2009 Aug;32(4):560-9. doi: 10.1007/s10545-009-1151-7. Epub 2009 Jun 20. J Inherit Metab Dis. 2009. PMID: 19544009 - SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes.
Grell TA, Goldman PJ, Drennan CL. Grell TA, et al. J Biol Chem. 2015 Feb 13;290(7):3964-71. doi: 10.1074/jbc.R114.581249. Epub 2014 Dec 4. J Biol Chem. 2015. PMID: 25477505 Free PMC article. Review. - Carbon extension in peptidylnucleoside biosynthesis by radical SAM enzymes.
Lilla EA, Yokoyama K. Lilla EA, et al. Nat Chem Biol. 2016 Nov;12(11):905-907. doi: 10.1038/nchembio.2187. Epub 2016 Sep 19. Nat Chem Biol. 2016. PMID: 27642865 Free PMC article.
References
- Reiss, J. (2000) Hum. Genet. 106, 157–163. - PubMed
- Reiss, J. & Johnson, J. L. (2003) Hum. Mutat. 21, 569–576. - PubMed
- Hänzelmann, P., Schwarz, G. & Mendel, R. R. (2002) J. Biol. Chem. 277, 18303–18312. - PubMed
- Hänzelmann, P., Hernández, H. L., Menzel, C., García-Serres, R., Huynh, B. H., Johnson, M. K., Mendel, R. R. & Schindelin, H. (2004) J. Biol. Chem. 279, 34721–34732. - PubMed
- Wuebbens, M. M. & Rajagopalan, K. V. (1995) J. Biol. Chem. 270, 1082–1087. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous