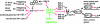Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans - PubMed (original) (raw)
Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans
Tao Liu et al. BMC Dev Biol. 2004.
Abstract
Background: When resources are scant, C. elegans larvae arrest as long-lived dauers under the control of insulin/IGF- and TGFbeta-related signaling pathways. However, critical questions remain regarding the regulation of this developmental event. How do three dozen insulin-like proteins regulate one tyrosine kinase receptor to control complex events in dauer, metabolism and aging? How are signals from the TGFbeta and insulin/IGF pathways integrated? What gene expression programs do these pathways regulate, and how do they control complex downstream events?
Results: We have identified genes that show different levels of expression in a comparison of wild-type L2 or L3 larvae (non-dauer) to TGFbeta mutants at similar developmental stages undergoing dauer formation. Many insulin/IGF pathway and other known dauer regulatory genes have changes in expression that suggest strong positive feedback by the TGFbeta pathway. In addition, many insulin-like ligand and novel genes with similarity to the extracellular domain of insulin/IGF receptors have altered expression. We have identified a large group of regulated genes with putative binding sites for the FOXO transcription factor, DAF-16. Genes with DAF-16 sites upstream of the transcription start site tend to be upregulated, whereas genes with DAF-16 sites downstream of the coding region tend to be downregulated. Finally, we also see strong regulation of many novel hedgehog- and patched-related genes, hormone biosynthetic genes, cell cycle genes, and other regulatory genes.
Conclusions: The feedback regulation of insulin/IGF pathway and other dauer genes that we observe would be predicted to amplify signals from the TGFbeta pathway; this amplification may serve to ensure a decisive choice between "dauer" and "non-dauer", even if environmental cues are ambiguous. Up and down regulation of insulin-like ligands and novel genes with similarity to the extracellular domain of insulin/IGF receptors suggests opposing roles for several members of these large gene families. Unlike in adults, most genes with putative DAF-16 binding sites are upregulated during dauer entry, suggesting that DAF-16 has different activity in dauer versus adult metabolism and aging. However, our observation that the position of putative DAF-16 binding sites is correlated with the direction of regulation suggests a novel method of achieving gene-specific regulation from a single pathway. We see evidence of TGFbeta-mediated regulation of several other classes of regulatory genes, and we discuss possible functions of these genes in dauer formation.
Figures
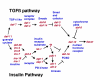
Figure 1
Signal transduction pathways that regulate dauer. Relationships between genes are based on mutant phenotypes and genetic interactions, gene expression in mutants, and homology to pathways in other organisms.
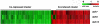
Figure 2
Cluster diagram of gene expression responses. Each row represents an average of 3–4 independent experiments. Each gene on the array is represented as a line in each column. The color of the line represents the log2 of the expression ratio, as indicated by the scale bar.
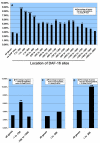
Figure 3
Regulation of genes with DAF-16 binding sites. In all panels, the bars show groups defined by the location of DAF-16 sites, as shown on the X-axis. We used overlapping intervals to allow robust statistical analysis. Negative numbers are bp upstream of the initiation codon, and positive numbers are bp downstream of the stop codon. Asterisks indicate statistical significance (exact hypergeometric probability) of the proportion of strongly regulated genes in the group compared to all genes:*** is p < 0.001, ** is p < 0.01, * is p < 0.05. A. Strongly upregulated (>2.1 fold, p < 0.01) genes. B. Two fold upregulated genes, excluding those genes in panel A. C. Strongly upregulated genes with downstream DAF-16 sites. D. Strongly downregulated (<2.1 fold, p < 0.01) genes.
Figure 4
Model for gene regulatory events under the control of TGFβ signaling. Transcriptional regulatory events suggested by expression data in this paper are indicated in red. The TGFβ pathway is shown in green; DAF-3 and DAF-5 form a transcription factor complex that function in neurons to control dauer entry [9]. DAF-3 and DAF-5 are shown regulating DAF-9 and DAF-12 indirectly because these genes are likely to be regulated in non-neuronal tissues. Insulin genes are shown as directly regulated by DAF-3 and DAF-5, but it is equally likely that these genes are regulated by feedback from the insulin pathway or by DAF-12.
Similar articles
- DAF-16-dependent and independent expression targets of DAF-2 insulin receptor-like pathway in Caenorhabditis elegans include FKBPs.
Yu H, Larsen PL. Yu H, et al. J Mol Biol. 2001 Dec 14;314(5):1017-28. doi: 10.1006/jmbi.2000.5210. J Mol Biol. 2001. PMID: 11743719 - pkc-1 regulates daf-2 insulin/IGF signalling-dependent control of dauer formation in Caenorhabditis elegans.
Monje JM, Brokate-Llanos AM, Pérez-Jiménez MM, Fidalgo MA, Muñoz MJ. Monje JM, et al. Aging Cell. 2011 Dec;10(6):1021-31. doi: 10.1111/j.1474-9726.2011.00747.x. Aging Cell. 2011. PMID: 21933341 - Lifespan and dauer regulation by tissue-specific activities of Caenorhabditis elegans DAF-18.
Masse I, Molin L, Billaud M, Solari F. Masse I, et al. Dev Biol. 2005 Oct 1;286(1):91-101. doi: 10.1016/j.ydbio.2005.07.010. Dev Biol. 2005. PMID: 16153634 - Alternate metabolism during the dauer stage of the nematode Caenorhabditis elegans.
Burnell AM, Houthoofd K, O'Hanlon K, Vanfleteren JR. Burnell AM, et al. Exp Gerontol. 2005 Nov;40(11):850-6. doi: 10.1016/j.exger.2005.09.006. Epub 2005 Oct 10. Exp Gerontol. 2005. PMID: 16221538 Review. - Worming pathways to and from DAF-16/FOXO.
Mukhopadhyay A, Oh SW, Tissenbaum HA. Mukhopadhyay A, et al. Exp Gerontol. 2006 Oct;41(10):928-34. doi: 10.1016/j.exger.2006.05.020. Epub 2006 Jul 12. Exp Gerontol. 2006. PMID: 16839734 Review.
Cited by
- Insulin signaling promotes germline proliferation in C. elegans.
Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Michaelson D, et al. Development. 2010 Feb;137(4):671-80. doi: 10.1242/dev.042523. Development. 2010. PMID: 20110332 Free PMC article. - The SR protein RSP-2 influences expression of the truncated insulin receptor DAF-2B in Caenorhabditis elegans.
Martinez BA, Gill MS. Martinez BA, et al. G3 (Bethesda). 2023 Jun 1;13(6):jkad064. doi: 10.1093/g3journal/jkad064. G3 (Bethesda). 2023. PMID: 36966398 Free PMC article. - Caenorhabditis elegans processes sensory information to choose between freeloading and self-defense strategies.
Schiffer JA, Servello FA, Heath WR, Amrit FRG, Stumbur SV, Eder M, Martin OM, Johnsen SB, Stanley JA, Tam H, Brennan SJ, McGowan NG, Vogelaar AL, Xu Y, Serkin WT, Ghazi A, Stroustrup N, Apfeld J. Schiffer JA, et al. Elife. 2020 May 5;9:e56186. doi: 10.7554/eLife.56186. Elife. 2020. PMID: 32367802 Free PMC article. - A Bystander Mechanism Explains the Specific Phenotype of a Broadly Expressed Misfolded Protein.
Klabonski L, Zha J, Senthilkumar L, Gidalevitz T. Klabonski L, et al. PLoS Genet. 2016 Dec 7;12(12):e1006450. doi: 10.1371/journal.pgen.1006450. eCollection 2016 Dec. PLoS Genet. 2016. PMID: 27926939 Free PMC article. - Reciprocal interactions between transforming growth factor beta signaling and collagens: Insights from Caenorhabditis elegans.
Goodman MB, Savage-Dunn C. Goodman MB, et al. Dev Dyn. 2022 Jan;251(1):47-60. doi: 10.1002/dvdy.423. Epub 2021 Sep 28. Dev Dyn. 2022. PMID: 34537996 Free PMC article.
References
- Riddle DL, Albert PS. Genetic and Environmental Regulation of dauer larva development. In: Riddle DL, Blumenthal T, Meyer BJ, Priess JR, editor. In C elegans II. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 739–768. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous