PhyloGenie: automated phylome generation and analysis - PubMed (original) (raw)
Comparative Study
. 2004 Sep 30;32(17):5231-8.
doi: 10.1093/nar/gkh867. Print 2004.
Affiliations
- PMID: 15459293
- PMCID: PMC521674
- DOI: 10.1093/nar/gkh867
Comparative Study
PhyloGenie: automated phylome generation and analysis
Tancred Frickey et al. Nucleic Acids Res. 2004.
Abstract
Phylogenetic reconstruction is the method of choice to determine the homologous relationships between sequences. Difficulties in producing high-quality alignments, which are the basis of good trees, and in automating the analysis of trees have unfortunately limited the use of phylogenetic reconstruction methods to individual genes or gene families. Due to the large number of sequences involved, phylogenetic analyses of proteomes preclude manual steps and therefore require a high degree of automation in sequence selection, alignment, phylogenetic inference and analysis of the resulting set of trees. We present a set of programs that automates the steps from seed sequence to phylogeny and a utility to extract all phylogenies that match specific topological constraints from a database of trees. Two example applications that show the type of questions that can be answered by phylome analysis are provided. The generation and analysis of the Thermoplasma acidophilum phylome with regard to lateral gene transfer between Thermoplasmata and Sulfolobus, showed best BLAST hits to be far less reliable indicators of lateral transfer than the corresponding protein phylogenies. The generation and analysis of the Danio rerio phylome provided more than twice as many proteins as described previously, supporting the hypothesis of an additional round of genome duplication in the actinopterygian lineage.
Figures
Figure 1
Alignment excerpts showing the most commonly encountered problems when converting BLAST or PSIBLAST HSPs to multiple alignments. (A) Three BLAST HSPs combined to a multiple sequence alignment and the resulting gapping problems. (B) Extreme examples of excessive and inconsistent gapping.
Figure 2
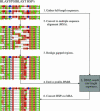
Layout showing the BLAST/PSIBLAST post-processing steps used to reduce excessive and inconsistent gapping. (1) All full-length sequences are gathered for HSPs and form the database used for HMM-searching in 5. (2) All HSPs matching E-value, score and coverage cutoff criteria are converted to a multiple sequence alignment. (3) The alignment sequences are filtered by maximum sequence identity to remove duplicate entries and gapped regions are realigned to resolve gapping problems. (4) A profile-HMM is derived from the multiple sequence alignment. (5) Sequences from step 1 are searched with the HMM generated in step 4 so as to better define the start and end of alignable regions and thereby improve alignment. (6) HMM-HSPs are converted to a multiple sequence alignment.
Figure 3
Tree rooting scheme. (a) Unrooted tree. (b) Tree rooted at the seed sequence (Man) with taxonomic “level” assignments for each node. (c) Tree rooted at the tipnode least related and most distant from the seed sequence (counting nodes) after the second round of taxonomic assignment. (d) Final tree, rooted at the most basal node the most distant from the seed sequence.
Figure 4
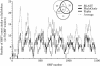
Chromosomal distribution of presumed laterally transferred ORFs between Thermoplasmata and Sulfolobus, according to PhyloGenie, Pyphy and best BLAST hits. The light gray, dark gray and black circles encompass the LGTs predicted by BLAST, Pyphy and PhyloGenie, respectively.
Similar articles
- Taxonomic colouring of phylogenetic trees of protein sequences.
Palidwor G, Reynaud EG, Andrade-Navarro MA. Palidwor G, et al. BMC Bioinformatics. 2006 Feb 17;7:79. doi: 10.1186/1471-2105-7-79. BMC Bioinformatics. 2006. PMID: 16503967 Free PMC article. - A database of phylogenetically atypical genes in archaeal and bacterial genomes, identified using the DarkHorse algorithm.
Podell S, Gaasterland T, Allen EE. Podell S, et al. BMC Bioinformatics. 2008 Oct 7;9:419. doi: 10.1186/1471-2105-9-419. BMC Bioinformatics. 2008. PMID: 18840280 Free PMC article. - Phylogenetic analyses: a brief introduction to methods and their application.
Horner DS, Pesole G. Horner DS, et al. Expert Rev Mol Diagn. 2004 May;4(3):339-50. doi: 10.1586/14737159.4.3.339. Expert Rev Mol Diagn. 2004. PMID: 15137901 Review. - Homology and phylogeny and their automated inference.
Fuellen G. Fuellen G. Naturwissenschaften. 2008 Jun;95(6):469-81. doi: 10.1007/s00114-008-0348-1. Epub 2008 Feb 21. Naturwissenschaften. 2008. PMID: 18288471 Review.
Cited by
- Plasmid-encoded toxin defence mediates mutualistic microbial interactions.
Moraïs S, Mazor M, Tovar-Herrera O, Zehavi T, Zorea A, Ifrach M, Bogumil D, Brandis A, Walter J, Elia N, Gur E, Mizrahi I. Moraïs S, et al. Nat Microbiol. 2024 Jan;9(1):108-119. doi: 10.1038/s41564-023-01521-9. Epub 2023 Dec 27. Nat Microbiol. 2024. PMID: 38151647 Free PMC article. - Modularity and diversity of target selectors in Tn7 transposons.
Faure G, Saito M, Benler S, Peng I, Wolf YI, Strecker J, Altae-Tran H, Neumann E, Li D, Makarova KS, Macrae RK, Koonin EV, Zhang F. Faure G, et al. Mol Cell. 2023 Jun 15;83(12):2122-2136.e10. doi: 10.1016/j.molcel.2023.05.013. Epub 2023 Jun 1. Mol Cell. 2023. PMID: 37267947 Free PMC article. - Reconstructing Yeasts Phylogenies and Ancestors from Whole Genome Data.
Feng B, Lin Y, Zhou L, Guo Y, Friedman R, Xia R, Hu F, Liu C, Tang J. Feng B, et al. Sci Rep. 2017 Nov 9;7(1):15209. doi: 10.1038/s41598-017-15484-5. Sci Rep. 2017. PMID: 29123238 Free PMC article. - Alienness: Rapid Detection of Candidate Horizontal Gene Transfers across the Tree of Life.
Rancurel C, Legrand L, Danchin EGJ. Rancurel C, et al. Genes (Basel). 2017 Sep 29;8(10):248. doi: 10.3390/genes8100248. Genes (Basel). 2017. PMID: 28961181 Free PMC article. - SICLE: a high-throughput tool for extracting evolutionary relationships from phylogenetic trees.
DeBlasio DF, Wisecaver JH. DeBlasio DF, et al. PeerJ. 2016 Aug 23;4:e2359. doi: 10.7717/peerj.2359. eCollection 2016. PeerJ. 2016. PMID: 27635331 Free PMC article.
References
- Koski L.B. and Golding,G.B. (2001) The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol., 52, 540–542. - PubMed
- International Human Genome Sequencing Consortium. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. - PubMed
- Stanhope M.J., Lupas,A.N., Italia,M.J., Koretke,K.K., Volker,C. and Brown,J.R. (2001) Phylogenetic analyses do not support horizontal gene transfers from bacteria to vertebrates. Nature, 411, 940–944. - PubMed
- Salzberg S.L., White,O., Peterson,J. and Eisen,J.A. (2001) Microbial genes in the human genome: lateral transfer or gene loss? Science, 5523, 1903–1906. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials