Centromere-encoded RNAs are integral components of the maize kinetochore - PubMed (original) (raw)
Centromere-encoded RNAs are integral components of the maize kinetochore
Christopher N Topp et al. Proc Natl Acad Sci U S A. 2004.
Abstract
RNA is involved in a variety of chromatin modification events, ranging from large-scale structural rearrangements to subtle local affects. Here, we extend the evidence for RNA-chromatin interactions to the centromere core. The data indicate that maize centromeric retrotransposons (CRMs) and satellite repeats (CentC) are not only transcribed, but that nearly half of the CRM and CentC RNA is tightly bound to centromeric histone H3 (CENH3), a key inner kinetochore protein. RNAs from another tandem repeat (180-bp knob sequence) or an abundant euchromatic retroelement (Opie) are undetectable within the same anti-CENH3 immune complexes. Both sense and antisense strands of CRM and CentC, but not small interfering RNAs homologous to either repeat, were found to coimmunoprecipitate with CENH3. The bulk of the immunoprecipitated RNA ranged in size from 40 to 200 nt. These data provide evidence for a pool of protected, single-stranded centromeric RNA within the centromere/kinetochore complex.
Figures
Fig. 2.
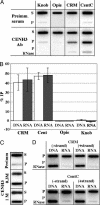
Centromere-encoded RNAs are coimmunoprecipitated with CENH3. (A) Chromatin samples were immunoprecipitated with anti-CENH3 antibodies, blotted for RNA, and probed with DNA probes for the sequences indicated. Supernatant (S) and pellet (P) fractions for the preimmune and anti-CENH3 treatments are shown. As shown in the bottom lane, RNaseA treatment removed the majority of RNA hybridization. These slot-blot images were used to acquire the numerical data in Table 1, experiment 1. (B) The RNA and DNA %IPs from five different experiments. The RNA %IP data (from Table 1) and associated DNA %IP values from each of five experiments are shown as mean ± SE. (C) High ionic strengths have a minimal impact on the immunoprecipitation of CentC RNA. The preimmmune (S and P) fractions are shown in the top two lanes, standard ChIP (using a 0.3 M NaCl wash) is shown in the middle two lanes, and standard ChIP after a 1 M NaCl wash is shown in the bottom two lanes. This experiment was not repeated in kind, but a second experiment with 0.7 M NaCl gave similar results. (D) Both strands of CRM and CentC are present after anti-CENH3-mediated ChIP. P fractions were blotted for the presence of DNA and RNA, either with or without prior RNaseA treatment. The sense and antisense strands are defined in Methods. This experiment was repeated three times with essentially identical results.
Fig. 3.
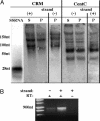
Centromeric siRNAs are not detected after CENH3-mediated ChIP. (A) The bulk of immunoprecipitated centromeric RNA ranges from 40 to 250 nt in length. The supernatant (S) and pellet (P) fractions are shown for both CRM and CentC. For the P fraction, hybridization with the opposite strand is shown. Hybridization to a 28-nt single-stranded synthetic RNA (SSRNA) homologous to the sense strand of the CRM GAG domain is shown at left. The 28-nt marker is underexposed relative to the other lanes. DNA markers (not shown, but indicated in nt) were used for higher molecular mass estimates. (B) Fragments (≈900 bp) of the CRM GAG domain were detected by strand-specific RT-PCR. This technique is not quantitative. The absence of product when no reverse transcriptase (RT–) was added indicates that the bands were derived from immunoprecipitated RNA, not DNA.
Fig. 1.
CRM is actively transcribed. (Upper) A map of CRM element (ref. , later called CRM2; ref. 31). The approximate locations of the GAG, reverse transcriptase (RT), and integrase (INT) domains are shown. (Lower) An agarose gel showing that several internal regions of CRM2 are readily amplified by RT-PCR. The locations of the amplified regions are indicated on the map.
Similar articles
- Centromeric retroelements and satellites interact with maize kinetochore protein CENH3.
Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, Nagaki K, Birchler JA, Jiang J, Dawe RK. Zhong CX, et al. Plant Cell. 2002 Nov;14(11):2825-36. doi: 10.1105/tpc.006106. Plant Cell. 2002. PMID: 12417704 Free PMC article. - Maize centromeres: organization and functional adaptation in the genetic background of oat.
Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, Dawe RK, Jiang J. Jin W, et al. Plant Cell. 2004 Mar;16(3):571-81. doi: 10.1105/tpc.018937. Epub 2004 Feb 18. Plant Cell. 2004. PMID: 14973167 Free PMC article. - Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize.
Liu Y, Su H, Zhang J, Liu Y, Feng C, Han F. Liu Y, et al. PLoS Biol. 2020 Jan 29;18(1):e3000582. doi: 10.1371/journal.pbio.3000582. eCollection 2020 Jan. PLoS Biol. 2020. PMID: 31995554 Free PMC article. - Centromeric chromatin and its dynamics in plants.
Lermontova I, Sandmann M, Mascher M, Schmit AC, Chabouté ME. Lermontova I, et al. Plant J. 2015 Jul;83(1):4-17. doi: 10.1111/tpj.12875. Plant J. 2015. PMID: 25976696 Review. - Complex structure of knobs and centromeric regions in maize chromosomes.
Ananiev EV, Phillips RL, Rines HW. Ananiev EV, et al. Tsitol Genet. 2000 Mar-Apr;34(2):11-5. Tsitol Genet. 2000. PMID: 10857197 Review.
Cited by
- 'Satellite DNA transcripts have diverse biological roles in Drosophila'.
Kuhn GC. Kuhn GC. Heredity (Edinb). 2015 Jul;115(1):1-2. doi: 10.1038/hdy.2015.12. Epub 2015 Mar 25. Heredity (Edinb). 2015. PMID: 25806543 Free PMC article. No abstract available. - Centromeric and pericentric transcription and transcripts: their intricate relationships, regulation, and functions.
Zhu J, Guo Q, Choi M, Liang Z, Yuen KWY. Zhu J, et al. Chromosoma. 2023 Sep;132(3):211-230. doi: 10.1007/s00412-023-00801-x. Epub 2023 Jul 4. Chromosoma. 2023. PMID: 37401943 Free PMC article. Review. - Using human artificial chromosomes to study centromere assembly and function.
Molina O, Kouprina N, Masumoto H, Larionov V, Earnshaw WC. Molina O, et al. Chromosoma. 2017 Oct;126(5):559-575. doi: 10.1007/s00412-017-0633-x. Epub 2017 Jul 7. Chromosoma. 2017. PMID: 28688039 Review. - α satellite DNA variation and function of the human centromere.
Sullivan LL, Chew K, Sullivan BA. Sullivan LL, et al. Nucleus. 2017 Jul 4;8(4):331-339. doi: 10.1080/19491034.2017.1308989. Epub 2017 Apr 13. Nucleus. 2017. PMID: 28406740 Free PMC article. - Functional Allium fistulosum Centromeres Comprise Arrays of a Long Satellite Repeat, Insertions of Retrotransposons and Chloroplast DNA.
Kirov I, Odintsov S, Omarov M, Gvaramiya S, Merkulov P, Dudnikov M, Ermolaev A, Van Laere K, Soloviev A, Khrustaleva L. Kirov I, et al. Front Plant Sci. 2020 Oct 23;11:562001. doi: 10.3389/fpls.2020.562001. eCollection 2020. Front Plant Sci. 2020. PMID: 33193489 Free PMC article.
References
- Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098–1102. - PubMed
- Amor, D. J., Kalitsis, P., Sumer, H. & Choo, K. H. A. (2004) Trends Cell Biol. 14, 359–368. - PubMed
- Choo, K. H. A. (2000) Trends Cell Biol. 10, 182–188. - PubMed
- Hooser, A. V., Ouspensik, I., Gregson, H., Starr, D., Yen, T., Goldberg, M., Yokomori, K., Earnshaw, W., Sullivan, K. & Brinkley, B. (2001) J. Cell Sci. 114, 3529–3542. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials