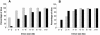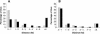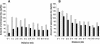A survey of RNA editing in human brain - PubMed (original) (raw)
Comparative Study
. 2004 Dec;14(12):2379-87.
doi: 10.1101/gr.2951204. Epub 2004 Nov 15.
Affiliations
- PMID: 15545495
- PMCID: PMC534661
- DOI: 10.1101/gr.2951204
Comparative Study
A survey of RNA editing in human brain
Matthew Blow et al. Genome Res. 2004 Dec.
Abstract
We have conducted a survey of RNA editing in human brain by comparing sequences of clones from a human brain cDNA library to the reference human genome sequence and to genomic DNA from the same individual. In the RNA sample from which the library was constructed, approximately 1:2000 nucleotides were edited out of >3 Mb surveyed. All edits were adenosine to inosine (A-->I) and were predominantly in intronic and in intergenic RNAs. No edits were found in translated exons and few in untranslated exons. Most edits were in high-copy-number repeats, usually Alus. Analysis of the genome in the vicinity of edited sequences strongly supports the idea that formation of intramolecular double-stranded RNA with an inverted copy underlies most A-->I editing. The likelihood of editing is increased by the presence of two inverted copies of a sequence within the same intron, proximity of the two sequences to each other (preferably within 2 kb), and by a high density of inverted copies in the vicinity. Editing exhibits sequence preferences and is less likely at an adenosine 3' to a guanosine and more likely at an adenosine 5' to a guanosine. Simulation by BLAST alignment of the double-stranded RNA molecules that underlie known edits indicates that there is a greater likelihood of A-->I editing at A:C mismatches than editing at other mismatches or at A:U matches. However, because A:U matches in double-stranded RNA are more common than all mismatches, overall the likely effect of editing is to increase the number of mismatches in double-stranded RNA.
Figures
Figure 1.
Proportion of edited and unedited _Alu_s with additional _Alu_s in the same intron. All _Alu_s aligning to the introns of known genes, and for which ≥80% of the genomic extent of the Alu was sequenced, were included in this analysis. The proportion of edited _Alu_s (gray bars) and the proportion of unedited _Alu_s (black bars) having an antisense Alu (A) or a same sense Alu (B) in the same intron are shown for different intron sizes.
Figure 2.
Distance from edited and unedited _Alu_s to the nearest Alu in the same intron. The Alu sequences included in this analysis are the same as in Figure 1. The proportion of edited _Alu_s (gray bars) and the proportion of unedited _Alu_s (black bars) at different distances from the nearest antisense Alu (A) or same sense Alu (B) are shown.
Figure 3.
Amount of flanking Alu sequence at different distances from edited and unedited _Alu_s. All _Alu_s for which ≥80% of the genomic extent of the Alu was sequenced were included in this analysis. For each Alu, the amount of flanking Alu sequence in the opposite orientation (A) or same orientation (B) in successive 1-kb windows was recorded. For each distance, the flanking Alu sequences in the 1-kb window 5′ and 3′ of the reference Alu were combined. The data presented are the average amount of Alu sequence flanking all edited _Alu_s (gray bars) or unedited _Alu_s (black bars).
Figure 4.
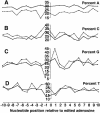
Sequence context of adenosines in edited Alu sequences. The sequence context of all edited adenosines and all unedited adenosines from all edited Alu sequences was compared. For each of the 10 bases either side of edited adenosines (dashed lines) and unedited adenosines (solid lines), the proportion of bases that were A, C, G, or T (_A_-D, respectively) at that position was calculated.
Figure 5.
Effect of sequence composition on the likelihood of RNA editing. A multiple alignment of all edited Alu sequences was prepared using CLUSTALW. At each position in the alignment, the proportion of edited adenosines was calculated from the number of sequenced edited adenosines and the total number of sequenced adenosines. The sequence composition at each position was calculated from all _Alu_s. For each position in the alignment, the proportion of edited adenosines is compared to the proportion of A, C, G, or T at that position (_A_-D, respectively).
Similar articles
- Identification of widespread ultra-edited human RNAs.
Carmi S, Borukhov I, Levanon EY. Carmi S, et al. PLoS Genet. 2011 Oct;7(10):e1002317. doi: 10.1371/journal.pgen.1002317. Epub 2011 Oct 20. PLoS Genet. 2011. PMID: 22028664 Free PMC article. - Detection of A-to-I Hyper-edited RNA Sequences.
Cohen-Fultheim R, Levanon EY. Cohen-Fultheim R, et al. Methods Mol Biol. 2021;2181:213-227. doi: 10.1007/978-1-0716-0787-9_13. Methods Mol Biol. 2021. PMID: 32729083 - Extensive adenosine-to-inosine editing detected in Alu repeats of antisense RNAs reveals scarcity of sense-antisense duplex formation.
Kawahara Y, Nishikura K. Kawahara Y, et al. FEBS Lett. 2006 Apr 17;580(9):2301-5. doi: 10.1016/j.febslet.2006.03.042. Epub 2006 Mar 24. FEBS Lett. 2006. PMID: 16574103 Free PMC article. - Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.
Levanon K, Eisenberg E, Rechavi G, Levanon EY. Levanon K, et al. EMBO Rep. 2005 Sep;6(9):831-5. doi: 10.1038/sj.embor.7400507. EMBO Rep. 2005. PMID: 16138094 Free PMC article. Review. - What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs.
Heraud-Farlow JE, Walkley CR. Heraud-Farlow JE, et al. Open Biol. 2020 Jul;10(7):200085. doi: 10.1098/rsob.200085. Epub 2020 Jul 1. Open Biol. 2020. PMID: 32603639 Free PMC article. Review.
Cited by
- Neurodegenerative diseases reflect the reciprocal roles played by retroelements in regulating memory and immunity.
Herbert A. Herbert A. Front Neurosci. 2024 Sep 20;18:1445540. doi: 10.3389/fnins.2024.1445540. eCollection 2024. Front Neurosci. 2024. PMID: 39371608 Free PMC article. - Increased RNA editing in maternal immune activation model of neurodevelopmental disease.
Tsivion-Visbord H, Kopel E, Feiglin A, Sofer T, Barzilay R, Ben-Zur T, Yaron O, Offen D, Levanon EY. Tsivion-Visbord H, et al. Nat Commun. 2020 Oct 16;11(1):5236. doi: 10.1038/s41467-020-19048-6. Nat Commun. 2020. PMID: 33067431 Free PMC article. - Transcriptome-wide identification of A > I RNA editing sites by inosine specific cleavage.
Cattenoz PB, Taft RJ, Westhof E, Mattick JS. Cattenoz PB, et al. RNA. 2013 Feb;19(2):257-70. doi: 10.1261/rna.036202.112. Epub 2012 Dec 21. RNA. 2013. PMID: 23264566 Free PMC article. - Identification of the long, edited dsRNAome of LPS-stimulated immune cells.
Blango MG, Bass BL. Blango MG, et al. Genome Res. 2016 Jun;26(6):852-62. doi: 10.1101/gr.203992.116. Epub 2016 Apr 21. Genome Res. 2016. PMID: 27197207 Free PMC article. - VIRGO: visualization of A-to-I RNA editing sites in genomic sequences.
Distefano R, Nigita G, Macca V, Laganà A, Giugno R, Pulvirenti A, Ferro A. Distefano R, et al. BMC Bioinformatics. 2013;14 Suppl 7(Suppl 7):S5. doi: 10.1186/1471-2105-14-S7-S5. Epub 2013 Apr 22. BMC Bioinformatics. 2013. PMID: 23815474 Free PMC article.
References
- Blanc, V. and Davidson, N.O. 2003. C-to-U RNA editing: Mechanisms leading to genetic diversity. J. Biol. Chem. 278: 1395-1398. - PubMed
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303-308. - PubMed
- Dawson, T.R., Sansam, C.L., and Emeson, R.B. 2004. Structure and sequence determinants required for the RNA editing of ADAR2 substrates. J. Biol. Chem. 279: 4941-4951. - PubMed
Web site references
- http://www.ensembl.org/; EnsEMBL.
- http://www.ncbi.nlm.nih.gov/SNP/; dbSNP.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous