Estimation of long-term effective population sizes through the history of durum wheat using microsatellite data - PubMed (original) (raw)
Estimation of long-term effective population sizes through the history of durum wheat using microsatellite data
A-C Thuillet et al. Genetics. 2005 Mar.
Abstract
Estimation of long-term effective population size (N(e)) from polymorphism data alone requires an independent knowledge of mutation rate. Microsatellites provide the opportunity to estimate N(e) because their high mutation rate can be estimated from observed mutations. We used this property to estimate N(e) in allotetraploid wheat Triticum turgidum at four stages of its history since its domestication. We estimated the mutation rate of 30 microsatellite loci. Allele-specific mutation rates mu were predicted from the number of repeats of the alleles. Effective population sizes were calculated from the diversity parameter theta = 4N(e)mu. We demonstrated from simulations that the unbiased estimator of theta based on Nei's heterozygosity is the most appropriate for estimating N(e) because of a small variance and a relative robustness to variations in the mutation model compared to other estimators. We found a N(e) of 32,500 individuals with a 95% confidence interval of [20,739; 45,991] in the wild ancestor of wheat, 12,000 ([5790; 19,300]) in the domesticated form, 6000 ([2831; 9556]) in landraces, and 1300 ([689; 2031]) in recent improved varieties. This decrease illustrates the successive bottlenecks in durum wheat. No selective effect was detected on our loci, despite a complete loss of polymorphism for two of them.
Figures
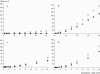
Figure 1.—
Magnitude of the bias on θ̂F (solid circles) and on θ̂_H_e (open circles) for different values of θ. The _x_-axis represents the parametric value of θ used for the simulations. Each point represents the average bias estimated on the basis of 1000 simulated data sets. Simulations were realized assuming either a constant population size and (a) a SMM or (b) a TPM or a bottlenecked population and (c) a SMM or (d) a TPM.
Figure 2.—
Correlation between θ̂F and the predicted mutation rate in the wild group of wheat (_R_2 = 0.56, P < 0.003).
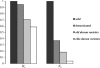
Figure 3.—
Reductions of heterozygosity (_H_e) and effective population size (_N_e) from the wild form, T. t. dicoccoides, to the elite varieties of durum wheat. Values are shown relative to the value in the wild form that is retained in the subsequent population.
Similar articles
- Microsatellite analysis reveals a progressive widening of the genetic basis in the elite durum wheat germplasm.
Maccaferri M, Sanguineti MC, Donini P, Tuberosa R. Maccaferri M, et al. Theor Appl Genet. 2003 Sep;107(5):783-97. doi: 10.1007/s00122-003-1319-8. Epub 2003 Jul 4. Theor Appl Genet. 2003. PMID: 12845433 - Genetic diversity and population structure analysis based on the high density SNP markers in Ethiopian durum wheat (Triticum turgidum ssp. durum).
Alemu A, Feyissa T, Letta T, Abeyo B. Alemu A, et al. BMC Genet. 2020 Feb 12;21(1):18. doi: 10.1186/s12863-020-0825-x. BMC Genet. 2020. PMID: 32050895 Free PMC article. - Direct estimation of mutation rate for 10 microsatellite loci in durum wheat, Triticum turgidum (L.) Thell. ssp durum desf.
Thuillet AC, Bru D, David J, Roumet P, Santoni S, Sourdille P, Bataillon T. Thuillet AC, et al. Mol Biol Evol. 2002 Jan;19(1):122-5. doi: 10.1093/oxfordjournals.molbev.a003977. Mol Biol Evol. 2002. PMID: 11752198 No abstract available. - Grinding up wheat: a massive loss of nucleotide diversity since domestication.
Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, Poncet C, Hochu I, Poirier S, Santoni S, Glémin S, David J. Haudry A, et al. Mol Biol Evol. 2007 Jul;24(7):1506-17. doi: 10.1093/molbev/msm077. Epub 2007 Apr 18. Mol Biol Evol. 2007. PMID: 17443011
Cited by
- Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data.
Bitocchi E, Nanni L, Bellucci E, Rossi M, Giardini A, Zeuli PS, Logozzo G, Stougaard J, McClean P, Attene G, Papa R. Bitocchi E, et al. Proc Natl Acad Sci U S A. 2012 Apr 3;109(14):E788-96. doi: 10.1073/pnas.1108973109. Epub 2012 Mar 5. Proc Natl Acad Sci U S A. 2012. PMID: 22393017 Free PMC article. - Chloroplast Microsatellite Diversity in Phaseolus vulgaris.
Desiderio F, Bitocchi E, Bellucci E, Rau D, Rodriguez M, Attene G, Papa R, Nanni L. Desiderio F, et al. Front Plant Sci. 2013 Jan 22;3:312. doi: 10.3389/fpls.2012.00312. eCollection 2012. Front Plant Sci. 2013. PMID: 23346091 Free PMC article. - Experimental estimation of mutation rates in a wheat population with a gene genealogy approach.
Raquin AL, Depaulis F, Lambert A, Galic N, Brabant P, Goldringer I. Raquin AL, et al. Genetics. 2008 Aug;179(4):2195-211. doi: 10.1534/genetics.107.071332. Epub 2008 Aug 9. Genetics. 2008. PMID: 18689900 Free PMC article. - Evidence for genetic differentiation and variable recombination rates among Dutch populations of the opportunistic human pathogen Aspergillus fumigatus.
Klaassen CH, Gibbons JG, Fedorova ND, Meis JF, Rokas A. Klaassen CH, et al. Mol Ecol. 2012 Jan;21(1):57-70. doi: 10.1111/j.1365-294X.2011.05364.x. Epub 2011 Nov 22. Mol Ecol. 2012. PMID: 22106836 Free PMC article. - Rapid evolution of simple sequence repeat induced by allopolyploidization.
Tang Z, Fu S, Ren Z, Zou Y. Tang Z, et al. J Mol Evol. 2009 Sep;69(3):217-28. doi: 10.1007/s00239-009-9261-2. Epub 2009 Aug 18. J Mol Evol. 2009. PMID: 19688286
References
- Brodehe, J., A. P. Møller and H. Ellegren, 2004. Individual variation in microsatellite mutation rate in barn swallows. Mutat. Res. 545: 73–80. - PubMed
- Buckler, IV, E. S., J. M. Thornsberry and S. Kresovich, 2001. Molecular diversity, structure and domestication of grasses. Genet. Res. 77: 213–218. - PubMed
- Casella, G., and R. L. Berger, 1990 Statistical Inference. Duxbury Press, Belmont, CA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials