Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays - PubMed (original) (raw)
. 2004 Sep-Oct;6(5):611-22.
doi: 10.1593/neo.04295.
Christian Pilarsky, Ole Ammerpohl, Jutta Lüttges, Armin Böhme, Bence Sipos, Melanie Foerder, Ingo Alldinger, Beatrix Jahnke, Hans Konrad Schackert, Holger Kalthoff, Bernd Kremer, Günter Klöppel, Hans Detlev Saeger
Affiliations
- PMID: 15548371
- PMCID: PMC1531666
- DOI: 10.1593/neo.04295
Gene expression profiling of microdissected pancreatic ductal carcinomas using high-density DNA microarrays
Robert Grützmann et al. Neoplasia. 2004 Sep-Oct.
Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains an important cause of malignancy-related death and is the eighth most common cancer with the lowest overall 5-year relative survival rate. To identify new molecular markers and candidates for new therapeutic regimens, we investigated the gene expression profile of microdissected cells from 11 normal pancreatic ducts, 14 samples of PDAC, and 4 well-characterized pancreatic cancer cell lines using the Affymetrix U133 GeneChip set. RNA was extracted from microdissected samples and cell lines, amplified, and labeled using a repetitive in vitro transcription protocol. Differentially expressed genes were identified using the significance analysis of microarrays program. We found 616 differentially expressed genes. Within these, 140 were also identified in PDAC by others, such as Galectin-1, Galectin-3, and MT-SP2. We validated the differential expression of several genes (e.g., CENPF, MCM2, MCM7, RAMP, IRAK1, and PTTG1) in PDAC by immunohistochemistry and reverse transcription polymerase chain reaction. We present a whole genome expression study of microdissected tissues from PDAC, from microdissected normal ductal pancreatic cells and pancreatic cancer cell lines using high-density microarrays. Within the panel of genes, we identified novel differentially expressed genes, which have not been associated with the pathogenesis of PDAC before.
Figures
Figure 1
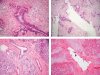
Manual microdissection of pancreatic tissue. Left: Before microdissection; right: after microdissection; upper panel: pancreatic ductal adenocarcinoma; lower panel: normal ductal epithelia.
Figure 2
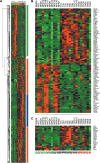
Analysis of gene expression in PDAC. (A) Hierarchical clustering of 14 microdissected PDACs, 11 microdissected normal ductal cells, and 4 established pancreatic tumor cell lines using the 616 differential gene set and a Euclidian distance matrix. The different colors reflect the predominant expression of the genes (green: normal tissue; blue: cell lines and PDAC; red: PDAC). The individual samples are colored (green: normal; blue: cell lines; red: PDAC). (B) Heat map of signature genes in PDAC. Genes were identified using the KNN method with a LOO validation step. (C) Heat map of protein kinase expression found in the set of differentially expressed genes. Genes that are upregulated appear in red, and those that are downregulated appear in green, with the expression value reflected by the intensity of the color.
Figure 3
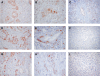
Immunohistochemical analysis of differentially expressed genes found in PDAC by microarray analysis. CENPF (A–C), MCM7 (D–F), and MCM2 (G–I) are highly expressed in the nucleus of PDACs of different grades (D: G1; A and E: G2; B, C, and F: G3; C, F, and I: normal pancreas). Original magnification, x100 (A, D, and E). Original magnification, x200 (C). Original magnification, x400 (B and F).
Figure 4
Validation of differentially expressed genes using RT-PCR. (A) RT-PCR analysis of five genes. RNA from tumor (T) and corresponding normal tissue (N) from four patients with a PDAC was isolated, and RT-PCR analysis for G6PD (control), PLAG1, PTTG, osf-2, and RAMP1 was performed. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. (B) Overview of RT-PCR results from normal pancreatic tissue, PDAC (upper panel), and cell lines (lower panel). GAPDH and G6PD expression analysis served as control. White background: not determined; green: no PCR signal (-), cT value > 35; yellow: PCR signal visible (-/+); orange: clear PCR signal (+), cT value > 27< 35; red: strong PCR signal (++), cT value > 20 < 27; dark red: very strong RT-PCR signal (+++), cT value < 20.
Similar articles
- Gene expression profiles of microdissected pancreatic ductal adenocarcinoma.
Grützmann R, Foerder M, Alldinger I, Staub E, Brümmendorf T, Röpcke S, Li X, Kristiansen G, Jesenofsky R, Sipos B, Löhr M, Lüttges J, Ockert D, Klöppel G, Saeger HD, Pilarsky C. Grützmann R, et al. Virchows Arch. 2003 Oct;443(4):508-17. doi: 10.1007/s00428-003-0884-1. Epub 2003 Aug 27. Virchows Arch. 2003. PMID: 12942322 - Small Nucleolar Noncoding RNA SNORA23, Up-Regulated in Human Pancreatic Ductal Adenocarcinoma, Regulates Expression of Spectrin Repeat-Containing Nuclear Envelope 2 to Promote Growth and Metastasis of Xenograft Tumors in Mice.
Cui L, Nakano K, Obchoei S, Setoguchi K, Matsumoto M, Yamamoto T, Obika S, Shimada K, Hiraoka N. Cui L, et al. Gastroenterology. 2017 Jul;153(1):292-306.e2. doi: 10.1053/j.gastro.2017.03.050. Epub 2017 Apr 5. Gastroenterology. 2017. PMID: 28390868 - Increased CDC20 expression is associated with pancreatic ductal adenocarcinoma differentiation and progression.
Chang DZ, Ma Y, Ji B, Liu Y, Hwu P, Abbruzzese JL, Logsdon C, Wang H. Chang DZ, et al. J Hematol Oncol. 2012 Apr 4;5:15. doi: 10.1186/1756-8722-5-15. J Hematol Oncol. 2012. PMID: 22475564 Free PMC article. - Critical role of laser microdissection for genetic, epigenetic and proteomic analyses in pancreatic cancer.
Funel N, Giovannetti E, Pollina LE, del Chiaro M, Mosca F, Boggi U, Campani D. Funel N, et al. Expert Rev Mol Diagn. 2011 Sep;11(7):695-701. doi: 10.1586/erm.11.62. Expert Rev Mol Diagn. 2011. PMID: 21902531 Review. - Microarray-based gene expression profiling in pancreatic ductal carcinoma: status quo and perspectives.
Grützmann R, Saeger HD, Lüttges J, Schackert HK, Kalthoff H, Klöppel G, Pilarsky C. Grützmann R, et al. Int J Colorectal Dis. 2004 Sep;19(5):401-13. doi: 10.1007/s00384-003-0563-3. Epub 2004 Jan 24. Int J Colorectal Dis. 2004. PMID: 14745573 Review.
Cited by
- Bioorthogonal labeling cell-surface proteins expressed in pancreatic cancer cells to identify potential diagnostic/therapeutic biomarkers.
Haun RS, Quick CM, Siegel ER, Raju I, Mackintosh SG, Tackett AJ. Haun RS, et al. Cancer Biol Ther. 2015;16(10):1557-65. doi: 10.1080/15384047.2015.1071740. Epub 2015 Jul 15. Cancer Biol Ther. 2015. PMID: 26176765 Free PMC article. - Complementary effects of HDAC inhibitor 4-PB on gap junction communication and cellular export mechanisms support restoration of chemosensitivity of PDAC cells.
Ammerpohl O, Trauzold A, Schniewind B, Griep U, Pilarsky C, Grutzmann R, Saeger HD, Janssen O, Sipos B, Kloppel G, Kalthoff H. Ammerpohl O, et al. Br J Cancer. 2007 Jan 15;96(1):73-81. doi: 10.1038/sj.bjc.6603511. Epub 2006 Dec 12. Br J Cancer. 2007. PMID: 17164759 Free PMC article. - Pancreatic satellite cells derived galectin-1 increase the progression and less survival of pancreatic ductal adenocarcinoma.
Tang D, Zhang J, Yuan Z, Gao J, Wang S, Ye N, Li P, Gao S, Miao Y, Wang D, Jiang K. Tang D, et al. PLoS One. 2014 Mar 4;9(3):e90476. doi: 10.1371/journal.pone.0090476. eCollection 2014. PLoS One. 2014. PMID: 24595374 Free PMC article. - The Immunohistochemical Expression of MCM-3, -5, and -7 Proteins in the Uterine Fibroids.
Rubisz P, Hirnle L, Kobierzycki C. Rubisz P, et al. Curr Issues Mol Biol. 2021 Jul 25;43(2):802-817. doi: 10.3390/cimb43020058. Curr Issues Mol Biol. 2021. PMID: 34449552 Free PMC article. - The G Protein-Coupled Receptor RAI3 Is an Independent Prognostic Factor for Pancreatic Cancer Survival and Regulates Proliferation via STAT3 Phosphorylation.
Jahny E, Yang H, Liu B, Jahnke B, Lademann F, Knösel T, Rümmele P, Grützmann R, Aust DE, Pilarsky C, Denz A. Jahny E, et al. PLoS One. 2017 Jan 23;12(1):e0170390. doi: 10.1371/journal.pone.0170390. eCollection 2017. PLoS One. 2017. PMID: 28114355 Free PMC article.
References
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics, 2003. CA Cancer J Clin. 2003;53:5–26. - PubMed
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. - PubMed
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, Kern SE. DPC4, a candidate tumor suppressor gene at human chromosome 18q21. Science. 1996;271:350–353. - PubMed
- Hruban RH, Offerhaus GJ, Kern SE, Goggins M, Wilentz RE, Yeo CJ. Tumor-suppressor genes in pancreatic cancer. J Hepatobiliary Pancr Surg. 1998;5:383–391. - PubMed
- Slebos RJ, Hoppin JA, Tolbert PE, Holly EA, Brock JW, Zhang RH, Bracci PM, Foley J, Stockton P, McGregor LM, Flake GP, Taylor JA. K-ras and p53 in pancreatic cancer: association with medical history, histopathology, and environmental exposures in a population-based study. Cancer Epidemiol Biomark Prev. 2000;9:1223–1232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous