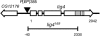End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent - PubMed (original) (raw)
End-joining repair of double-strand breaks in Drosophila melanogaster is largely DNA ligase IV independent
Mitch McVey et al. Genetics. 2004 Dec.
Abstract
Repair of DNA double-strand breaks can occur by either nonhomologous end joining or homologous recombination. Most nonhomologous end joining requires a specialized ligase, DNA ligase IV (Lig4). In Drosophila melanogaster, double-strand breaks created by excision of a P element are usually repaired by a homologous recombination pathway called synthesis-dependent strand annealing (SDSA). SDSA requires strand invasion mediated by DmRad51, the product of the spn-A gene. In spn-A mutants, repair proceeds through a nonconservative pathway involving the annealing of microhomologies found within the 17-nt overhangs produced by P excision. We report here that end joining of P-element breaks in the absence of DmRad51 does not require Drosophila LIG4. In wild-type flies, SDSA is sometimes incomplete, and repair is finished by an end-joining pathway that also appears to be independent of LIG4. Loss of LIG4 does not increase sensitivity to ionizing radiation in late-stage larvae, but lig4 spn-A double mutants do show heightened sensitivity relative to spn-A single mutants. Together, our results suggest that a LIG4-independent end-joining pathway is responsible for the majority of double-strand break repair in the absence of homologous recombination in flies.
Figures
Figure 1.—
The Lig4 region and the lig4169 deletion. The four coding exons for LIG4 are shown as boxes; the shaded box represents the BRCT motif, and the stippled box corresponds to the region responsible for binding CG3448, the putative Drosophila XRCC4 homolog. Transcription of the adjacent CG12176 locus occurs in the opposite direction of Lig4. The P{EP}385 element used to create the lig4169 deletion is located 40 bp upstream of Lig4. The lig4169 deletion extends from −40 to +2330 (+1 corresponds to the translation start site).
Figure 2.—
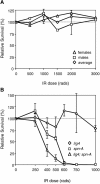
lig4169 mutants are not sensitive to ionizing radiation. (A) Third-instar larvae from the cross lig4169 /+ × lig4169/Y were irradiated with indicated doses of gamma rays, and survival rates of lig4169/lig4169 homozygous to lig4169/+ heterozygous female progeny, or lig4169/Y to +/Y male progeny, were calculated. Survival rates are given relative to the ratios observed for the untreated controls. (▵) females, (□) males, (⋄) average. (B) Mutation of lig4 and spn-A results in synergistic sensitivity to intermediate doses of X rays. Double-mutant flies were created by crossing lig4169/FM7; spn-A57/TM3 females to lig4169/Y; spn-A93/TM3 males. Third-instar larvae were exposed to indicated doses of gamma rays and relative survival rates were calculated relative to the survival rate of lig4 spn-A heterozygous females at each dose. (⋄) lig4, (□) spn-A, (▵) lig4 spn-A. Heightened sensitivity was observed in the double mutant at doses between 400 and 500 rad.
Figure 3.—
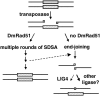
Model for repair of _P_-element-induced DSBs in Drosophila. Transposase cuts at both ends of the P element, creating a double-strand gap with 3′ overhanging ends that can invade into the unbroken sister chromatid (shaded) and initiate homologous repair by SDSA. Occasionally, for unknown reasons, SDSA aborts and repair of the broken chromosome occurs by end joining (dashed line). In the absence of the strand-exchange protein DmRad51, SDSA cannot initiate and end joining proceeds by alignment of small microhomologies, presumably followed by trimming of overhanging ends, filling in of gaps, and ligation. The majority of this end-joining repair occurs via a LIG4-independent pathway.
Similar articles
- Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila.
Bozas A, Beumer KJ, Trautman JK, Carroll D. Bozas A, et al. Genetics. 2009 Jul;182(3):641-51. doi: 10.1534/genetics.109.101329. Epub 2009 Apr 20. Genetics. 2009. PMID: 19380480 Free PMC article. - The Drosophila melanogaster DNA Ligase IV gene plays a crucial role in the repair of radiation-induced DNA double-strand breaks and acts synergistically with Rad54.
Gorski MM, Eeken JC, de Jong AW, Klink I, Loos M, Romeijn RJ, van Veen BL, Mullenders LH, Ferro W, Pastink A. Gorski MM, et al. Genetics. 2003 Dec;165(4):1929-41. doi: 10.1093/genetics/165.4.1929. Genetics. 2003. PMID: 14704177 Free PMC article. - DNA ligase IV mutations confer shorter lifespan and increased sensitivity to nutrient stress in Drosophila melanogaster.
Joshi R, Banerjee SJ, Curtiss J, Ashley AK. Joshi R, et al. J Appl Genet. 2022 Feb;63(1):141-144. doi: 10.1007/s13353-021-00637-0. Epub 2021 Nov 24. J Appl Genet. 2022. PMID: 34817771 Free PMC article. - Mammalian DNA ligases; roles in maintaining genome integrity.
Sallmyr A, Bhandari SK, Naila T, Tomkinson AE. Sallmyr A, et al. J Mol Biol. 2024 Jan 1;436(1):168276. doi: 10.1016/j.jmb.2023.168276. Epub 2023 Sep 13. J Mol Biol. 2024. PMID: 37714297 Free PMC article. Review. - Structure and function of mammalian DNA ligases.
Tomkinson AE, Mackey ZB. Tomkinson AE, et al. Mutat Res. 1998 Feb;407(1):1-9. doi: 10.1016/s0921-8777(97)00050-5. Mutat Res. 1998. PMID: 9539976 Review.
Cited by
- CRISPR/Cas-based precision genome editing via microhomology-mediated end joining.
Van Vu T, Thi Hai Doan D, Kim J, Sung YW, Thi Tran M, Song YJ, Das S, Kim JY. Van Vu T, et al. Plant Biotechnol J. 2021 Feb;19(2):230-239. doi: 10.1111/pbi.13490. Epub 2020 Nov 9. Plant Biotechnol J. 2021. PMID: 33047464 Free PMC article. Review. - Identification of a micropeptide and multiple secondary cell genes that modulate Drosophila male reproductive success.
Immarigeon C, Frei Y, Delbare SYN, Gligorov D, Machado Almeida P, Grey J, Fabbro L, Nagoshi E, Billeter JC, Wolfner MF, Karch F, Maeda RK. Immarigeon C, et al. Proc Natl Acad Sci U S A. 2021 Apr 13;118(15):e2001897118. doi: 10.1073/pnas.2001897118. Proc Natl Acad Sci U S A. 2021. PMID: 33876742 Free PMC article. - Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells.
Böttcher R, Hollmann M, Merk K, Nitschko V, Obermaier C, Philippou-Massier J, Wieland I, Gaul U, Förstemann K. Böttcher R, et al. Nucleic Acids Res. 2014 Jun;42(11):e89. doi: 10.1093/nar/gku289. Epub 2014 Apr 19. Nucleic Acids Res. 2014. PMID: 24748663 Free PMC article. - Whole-Genome Analysis of Individual Meiotic Events in Drosophila melanogaster Reveals That Noncrossover Gene Conversions Are Insensitive to Interference and the Centromere Effect.
Miller DE, Smith CB, Kazemi NY, Cockrell AJ, Arvanitakis AV, Blumenstiel JP, Jaspersen SL, Hawley RS. Miller DE, et al. Genetics. 2016 May;203(1):159-71. doi: 10.1534/genetics.115.186486. Epub 2016 Mar 4. Genetics. 2016. PMID: 26944917 Free PMC article. - A pathway for error-free non-homologous end joining of resected meiotic double-strand breaks.
Hatkevich T, Miller DE, Turcotte CA, Miller MC, Sekelsky J. Hatkevich T, et al. Nucleic Acids Res. 2021 Jan 25;49(2):879-890. doi: 10.1093/nar/gkaa1205. Nucleic Acids Res. 2021. PMID: 33406239 Free PMC article.
References
- Adams, M. D., M. McVey and J. J. Sekelsky, 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. - PubMed
- Barnes, D. E., G. Stamp, I. Rosewell, A. Denzel and T. Lindahl, 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 8: 1395–1398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K12 GM000678/GM/NIGMS NIH HHS/United States
- R01 GM061252/GM/NIGMS NIH HHS/United States
- GM-000678/GM/NIGMS NIH HHS/United States
- R01 GM-61252/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials