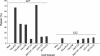Short-patch correction of C/C mismatches in human cells - PubMed (original) (raw)
. 2004 Dec 21;32(22):6696-705.
doi: 10.1093/nar/gkh990. Print 2004.
Affiliations
- PMID: 15613598
- PMCID: PMC545458
- DOI: 10.1093/nar/gkh990
Short-patch correction of C/C mismatches in human cells
Regula Muheim-Lenz et al. Nucleic Acids Res. 2004.
Abstract
We examined whether the human nucleotide excision repair complex, which is specialized on the removal of bulky DNA adducts, also displays a correcting activity on base mismatches. The cytosine/cytosine (C/C) lesion was used as a model substrate to monitor the correction of base mismatches in human cells. Fibroblasts with different repair capabilities were transfected with shuttle vectors that contain a site-directed C/C mismatch in the replication origin, accompanied by an additional C/C mismatch in one of the flanking sequences that are not essential for replication. Analysis of the vector progeny obtained from these doubly modified substrates revealed that C/C mismatches were eliminated before DNA synthesis not only in the repair-proficient background, but also when the target cells carried a genetic defect in long-patch mismatch repair, in nucleotide excision repair, or when both pathways were deleted. Furthermore, cells deficient for long-patch mismatch repair as well as a cell line that combines mismatch and nucleotide excision repair defects were able to correct multiple C/C mispairs, placed at distances of 21-44 nt, in an independent manner, such that the removal of each lesion led to individual repair patches. These results support the existence of a concurrent short-patch mechanism that rectifies C/C mismatches.
Figures
Figure 1
Assay for in vitro human NER activity. Internally 32P-labeled linear substrates of 147 bp were incubated with HeLa whole cell extract. The substrate contained various mismatches as indicated (lanes 2–6), a site-specific AAF-dG adduct (lane 8) or a site-specific pivaloyl backbone adduct (lanes 9) in the central BstBI sequence. UM, unmodified homoduplex substrate. The gel has been overexposed to demonstrate the absence of detectable activity in response to the mismatches.
Figure 2
Excision of mismatches in an in vitro assay for human MMR activity. Heteroduplex bacteriophage substrates, containing a G/T or C/C mismatch, were incubated with cytoplamic extracts (CE), whole cell extracts (WCE) or nuclear extracts (NE) prepared from different cell lines. MMR efficiency was quantified after transformation of an E.coli mutS strain with the in vitro repair products. The mock-treated sample resulted from an incubation without cell extract. Complementation was performed by the addition of hMutLα to 293T cytoplasmic extracts. Lovo and HCT15 are human cell lines deficient for hMSH2 and hMSH6, respectively.
Figure 3
Vector pS189 derivatives containing two C/C mismatches. (A) Scheme illustrating the location of C/CSfiI (position 3578) in the SV40 origin of pS189. The pentanucleotide recognition sequences for T antigen binding are indicated by the arrows. (B) Composition of substrates 1 and 2. (C) Restriction analysis of substrate 1 (lanes 4–6) and substrate 2 (lanes 9–11). The homoduplex vector was prepared by extension and ligation of unmodified primers. Control, no restriction endonuclease added.
Figure 4
Independent processing of two neighboring C/C mismatches in human fibroblasts. (A) SfiI and Acc65I restriction analysis of clones 1–4 isolated after transfection of substrate 1 in wild-type fibroblasts. Control, no enzyme added. (B) Schematic view of the two different repair products isolated after transfection of substrate 1. (C) AvrII restriction analysis of clones number 1–14 isolated after transfection of substrate 2 in wild-type fibroblasts. (D) Scheme of the two different repair products of substrate 2.
Figure 5
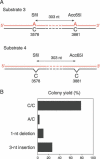
Processing of different lesions in long-patch MMR-deficient 293T cells. (A) Scheme of substrate 3 (pS189 containing two neighboring A/C mismatches) and substrate 4 (pS189 containing two neighboring single nucleotide deletions). (B) Relative yield of progeny after transfection of pS189 derivatives, carrying the indicated lesions, in 293T cells. Colony numbers (mean values of two independent experiments) are expressed as the percentages of bacterial colonies obtained after transfection of the same cells with unmodified control vector.
Figure 6
Repair of C/C mismatches in long-patch MMR-deficient 293T cells. (A) SfiI and Acc65I restriction analysis of clones 1–4 isolated after transfection of substrate 1 in 293T cells. (B) Restriction analysis by AvrII of clones 1–20 isolated after transfection of substrate 2 in 293T cells.
Figure 7
Short-patch repair of C/C mismatches. (A) Schematic view of substrate 6 containing three closely spaced C/C mismatches. (B) Sequence of a representative clone isolated after transfection of substrate 6 in MMR- and NER-defective XP12ROB4 fibroblasts. Red arrows indicate the original position of C/C mismatches in the substrate.
Similar articles
- Discrimination and versatility in mismatch repair.
Hays JB, Hoffman PD, Wang H. Hays JB, et al. DNA Repair (Amst). 2005 Dec 8;4(12):1463-74. doi: 10.1016/j.dnarep.2005.09.002. Epub 2005 Oct 5. DNA Repair (Amst). 2005. PMID: 16213799 - Characterization of palindromic loop mismatch repair tracts in mammalian cells.
Miller CA, Bill CA, Nickoloff JA. Miller CA, et al. DNA Repair (Amst). 2004 Apr 1;3(4):421-8. doi: 10.1016/j.dnarep.2003.12.006. DNA Repair (Amst). 2004. PMID: 15010318 - The rate of base excision repair of uracil is controlled by the initiating glycosylase.
Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE. Visnes T, et al. DNA Repair (Amst). 2008 Nov 1;7(11):1869-81. doi: 10.1016/j.dnarep.2008.07.012. Epub 2008 Sep 4. DNA Repair (Amst). 2008. PMID: 18721906 - [Mismatch repair].
Goliasnaia NV, Tsvetkova NA. Goliasnaia NV, et al. Mol Biol (Mosk). 2006 Mar-Apr;40(2):211-23. Mol Biol (Mosk). 2006. PMID: 16637261 Review. Russian. - [Eukaryotic error prone DNA polymerases: suggested roles in replication, repair and mutagenesis].
Krutiakov VM. Krutiakov VM. Mol Biol (Mosk). 2006 Jan-Feb;40(1):3-11. Mol Biol (Mosk). 2006. PMID: 16523685 Review. Russian.
Cited by
- Alleviation of C⋅C Mismatches in DNA by the Escherichia coli Fpg Protein.
Tesfahun AN, Alexeeva M, Tomkuvienė M, Arshad A, Guragain P, Klungland A, Klimašauskas S, Ruoff P, Bjelland S. Tesfahun AN, et al. Front Microbiol. 2021 Jun 30;12:608839. doi: 10.3389/fmicb.2021.608839. eCollection 2021. Front Microbiol. 2021. PMID: 34276575 Free PMC article. - Eliminating both canonical and short-patch mismatch repair in Drosophila melanogaster suggests a new meiotic recombination model.
Crown KN, McMahan S, Sekelsky J. Crown KN, et al. PLoS Genet. 2014 Sep 4;10(9):e1004583. doi: 10.1371/journal.pgen.1004583. eCollection 2014 Sep. PLoS Genet. 2014. PMID: 25188408 Free PMC article. - Removal of deoxyinosine from the Escherichia coli chromosome as studied by oligonucleotide transformation.
Weiss B. Weiss B. DNA Repair (Amst). 2008 Feb 1;7(2):205-12. doi: 10.1016/j.dnarep.2007.09.010. Epub 2007 Nov 5. DNA Repair (Amst). 2008. PMID: 17981100 Free PMC article. - Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2.
Smith JA, Bannister LA, Bhattacharjee V, Wang Y, Waldman BC, Waldman AS. Smith JA, et al. Mol Cell Biol. 2007 Nov;27(22):7816-27. doi: 10.1128/MCB.00455-07. Epub 2007 Sep 10. Mol Cell Biol. 2007. PMID: 17846123 Free PMC article. - Trypanosoma brucei homologous recombination is dependent on substrate length and homology, though displays a differential dependence on mismatch repair as substrate length decreases.
Barnes RL, McCulloch R. Barnes RL, et al. Nucleic Acids Res. 2007;35(10):3478-93. doi: 10.1093/nar/gkm249. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478508 Free PMC article.
References
- Kolodner R. (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev., 10, 1433–1442. - PubMed
- Schär P. and Jiricny,J. (1998) Eukaryotic mismatch repair. In Eckstein,F.A. and David,M.J. (eds), Nucleic Acids and Molecular Biology. Springer, NY, Vol. XII, pp. 199–246.
- Marti T.M., Kunz,C. and Fleck,O. (2002) DNA mismatch repair and mutation avoidance pathways. J. Cell. Physiol., 191, 28–41. - PubMed
- Modrich P. and Lahue,R. (1996) Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem., 65, 101–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials