Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing - PubMed (original) (raw)
Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing
Chryssa Kanellopoulou et al. Genes Dev. 2005.
Abstract
Dicer is the enzyme that cleaves double-stranded RNA (dsRNA) into 21-25-nt-long species responsible for sequence-specific RNA-induced gene silencing at the transcriptional, post-transcriptional, or translational level. We disrupted the dicer-1 (dcr-1) gene in mouse embryonic stem (ES) cells by conditional gene targeting and generated Dicer-null ES cells. These cells were viable, despite being completely defective in RNA interference (RNAi) and the generation of microRNAs (miRNAs). However, the mutant ES cells displayed severe defects in differentiation both in vitro and in vivo. Epigenetic silencing of centromeric repeat sequences and the expression of homologous small dsRNAs were markedly reduced. Re-expression of Dicer in the knockout cells rescued these phenotypes. Our data suggest that Dicer participates in multiple, fundamental biological processes in a mammalian organism, ranging from stem cell differentiation to the maintenance of centromeric heterochromatin structure and centromeric silencing.
Figures
Figure 1.
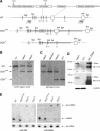
Conditional gene targeting of mouse dcr-1. (A) Depicted at the top is the structure of the Dicer protein composed of an N-terminal RNA helicase domain, PAZ domain, followed by two catalytic ribonuclease III domains and a double-stranded RNA-binding domain. Epitopes used for generation of anti-Dicer antibodies are indicated. Below it, part of the wild-type (WT) dcr-1 gene is depicted. Exons 18–20, encoding half of the PAZ domain and the first RNase III domain, are flanked by loxP sites (triangles). The locations of BamHI (BHI) and BglI restriction sites and 5′ and 3′ probes used for screening of homologous recombinants are indicated. Homologous recombination of our targeting vector (bold line) resulted in the allele depicted as DCRneo. An _FRT_-flanked PGK-neomycin cassette (neo†) is inserted into intron 17. Upon Cre-mediated recombination of the introduced loxP sites, a Dcr-null allele is generated (depicted as DCRΔ). (B) Southern blot of BamHI-digested genomic DNA from a wild-type (+/+) ES cell clone, and two homologous recombinants (neo/+) were hybridized with the 3′ probe (see A). (C) Southern blot of BglI-digested genomic DNA from a heterozygous (neo/+) ES cell clone, a homozygous (neo/neo) clone, a heterozygous deleted clone (neo/Δ), and a homozygous (Δ/Δ) Dcr-null clone was hybridized with the 5′ probe (see A). (D) Western blot of extracts derived from a heterozygous (neo/+) ES cell clone, a homozygous (Δ/Δ) clone, and recombinant Dicer (rec. Dicer) protein. The Western blot was probed for Dicer protein using antisera raised against two different peptides homologous to mouse Dicer (the corresponding epitopes, 1414 and 1416, are indicated on the schematic representation of Dicer protein, A), and then stripped and reprobed for tubulin (bottom). (E) Northern analysis of miRNA expression in mouse ES cells. Total RNA from a control wild-type and Dicer-proficient clone (neo/neo) and two Dcr-null clones (Δ/Δ) was resolved in a denaturing polyacrylamide gel and transferred onto a nylon membrane. Shown are PhosphorImages of two blots hybridized either to a radiolabeled oligonucleotide complementary to miR-293 (left panel) or miR-292as (right panel). To control for equal RNA loading, the Northern blots were also reprobed for leucine transfer RNA (Leu tRNA). The arrows indicate Leu tRNA, pre-miRNA, and processed mature miRNA.
Figure 2.
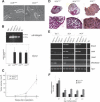
Dicer-deficient ES cells retain ES cell characteristics but fail to differentiate. (A) Phase-contrast micrographs of heterozygous (neo/+) and homozygous (Δ/Δ)ES cell colonies (indicated by arrows) grown on a feeder layer of MEFs. (B, top panel) RT–PCR analysis of the ES cell-specific α6-integrin isoform (306 bp). (Bottom panel) Quantitative RT–PCR analysis of oct-4 transcripts normalized to hprt transcripts. The averages and standard deviations of three experiments are depicted in the graph. (C) Size of teratomas following injection of the indicated ES cells into nude mice is plotted as a function of time (weeks post-implantation). The averages from five injected mice are shown. (D) Light micrographs of paraffin sections from day 12 EBs stained with hematoxylin-eosin. EBs derived from heterozygous (DCRneo/+), homozygous (DCRneo/neo), and Dicer-deficient ES cells (DCRΔ/Δ) are shown in the top panel (10×). In the lower panel, higher magnification images (40×) of a portion of the same EBs are displayed. (E) Semiquantitative RT–PCR analyses of mesoderm- and ectoderm-specific differentiation markers. RNA was extracted and reverse-transcribed from EBs generated from DCRneo/+, DCRneo/neo, and two different DCRΔ/Δ clones at day 3, 5, and 8 of differentiation and analyzed for expression of differentiation markers bmp4, hnf4, gata1, and brachyury. Hprt transcripts were amplified as a loading control. (F) Quantitative real-time PCR analysis of oct-4 transcripts normalized to hprt transcripts in EBs at days 3, 5, 8, 10, and12 of differentiation. The averages and standard deviations of three experiments are depicted in the graph.
Figure 3.
Analyses of satellite repeat transcripts in ES cells in the presence or absence of Dicer. (A) Quantitative real-time RT–PCR was performed to determine the relative abundance of transcripts from pericentric major satellite repeats in control (Dcrneo/+, gray bars) and mutant (DcrΔ/Δ, black bars) ES cells. Total RNA samples were treated with DNase I prior to reverse transcription. Plotted are values (in arbitrary units) for abundance of major satellite transcripts normalized to levels of hprt transcripts, a housekeeping gene. ES cells were either untreated or treated with increasing concentrations of the histone deactylase inhibitor, TSA. (B) Semiquantitative RT–PCR was performed to determine the relative abundance of transcripts from centromeric minor satellite repeats in control (neo/+ and neo/neo) and mutant (Δ/Δ) ES cells. Digital photographs (negative image) of agarose gel analyses of the RT–PCR products for minor satellite and β_-actin_ transcripts are shown. Total RNA samples were treated with DNase I prior to reverse transcription. Equivalent amounts of reverse-transcribed RNA were used for each PCR reaction as determined by the abundance of hprt transcripts, which was obtained by quantitative real-time RT–PCR (data not shown). (C) Total RNA from wild-type ES cells was either untreated (–) or treated (+) with RNase ONE and resolved in a denaturing 15% acrylamide gel. Shown in the left panel is the gel stained with ethidium bromide (EtBr). In the right panel, the gel was blotted and probed with a radiolabeled probe specific for minor satellite repeats (pMR150). A radiolabeled Decade marker (M) consisting of 20-, 30-, 40-, 50-, 60-, 70-, 80-, 90-, 100-, and 150-nt RNA species is included. (D) Total RNA was size fractionated using mirVana glass fiber filters and equivalent amounts of the small RNA fraction (<200 nt) were resolved in a denaturing 15% acrylamide gel. Shown in the _left_ panel is the gel stained with EtBr loaded with short RNAs from wild-type (neo/+) or mutant (Δ/Δ) ES cells. In the _right_ panel, the gel was used for a Northern blot hybridized with a probe specific for minor satellite repeats (pMR150). A radiolabeled RNA Decade ladder (M) is also included. (_E_) Total RNA was size fractionated using mirVana glass fiber filters, and equivalent amounts of the long RNA fraction (>200 nt) were resolved in a denaturing 10% acrylamide gel and analyzed by Northern blot as in C and D. The EtBr-stained gel is shown below.
Figure 4.
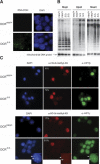
Evidence for transcriptional derepression of centromeric repeats. (A) RNA FISH using a centromeric DNA probe was performed on DCRneo/+ and DCRΔ/Δ ES cells. The respective DAPI nuclear staining is shown next to each panel (63×). (B) DNA methylation analysis of genomic DNA derived from two DCRneo/+ and two DCRΔ/Δ clones. Equivalent amounts of DNA were digested with MspI, HpaII, or MaeII. (Lower panel) EtBr staining of the gel (data not shown) and rehybridization with a mitochondrial DNA probe demonstrate equivalent loading. MspI cleaves irrespective of whether the CpG dinucleotide within the cognate restriction site is methylated, whereas HpaII or MaeII will not. Gels were blotted on a nitrocellulose membrane and hybridized with a probe specific for the minor satellite repeat. (C) Histone modifications and HP1 protein localization are altered in Dicer-deficient cells. DCRneo/+ and DCRΔ/Δ cells were stained either with a rabbit polyclonal antibody specific for dimethyl-H3K9 and a monoclonal antibody against HP1β (upper panels) or a polyclonal antibody specific for trimethyl-H3K9 and a monoclonal antibody against HP1γ (lower panels). DAPI staining of nuclei for each panel is shown on the left (63× magnification). Percentages of cells unstained or dim for variously methylated H3K9 species are indicated on the top left corner of each panel. At least 200 cells from each clone were counted. Inset figures point to an example of the colocalization of HP-1γ, trimethyl H3K9 staining, and centromeric heterochromatin as reflected by the existence of DAPI dense spots.
Figure 5.
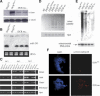
Reconstitution of Dicer expression and function in DCRΔ/Δ ES cells. (A) Western blot of extracts prepared from a homozygous (Δ/Δ) clone, a heterozygous (neo/+) ES cell clone, and two independent Dicer-reconstituted clones (DCR rec.). (Bottom) The Western blot was probed for Dicer protein using antisera (ab1414) and then stripped and reprobed for tubulin. (B) Northern analysis of miR-293 expression in total RNA from a control ES cell clone (neo/+), a Dcr-null clone (Δ/Δ), and Dicer-reconstituted clones performed as in Figure 1E. (Bottom) To demonstrate equal RNA loading, a segment of the EtBr-stained gel is shown. (C) Semiquantitative RT–PCR analyses of mesoderm- and ectoderm-specific differentiation markers. RNA was extracted and reverse-transcribed from EBs generated from DCRneo/+, DCRneo/neo, and two different DCRΔ/Δ clones at day 3, 5, and 8 of differentiation and analyzed for expression of differentiation markers bmp4, hnf4, gata1, and brachyury. Hprt transcripts were amplified as a loading control. (D) Semiquantitative RT–PCR of transcripts from centromeric minor satellite repeats in control (DCRneo/+ and DCRneo/neo), two mutants (DCRΔ/Δ), and Dicer-reconstituted (DCR rec.) ES cells. Digital photographs (negative image) of agarose gel analyses of the RT–PCR products for minor satellite and hprt transcripts are shown. Total RNA samples were treated with DNase I prior to reverse transcription. (E) DNA methylation analysis of genomic DNA derived from DCRneo/+, two DCRΔ/Δ, and two Dicer-reconstituted (DCR rec.) clones. Equivalent amounts of DNA were digested with HpaII and resolved in a 1% agarose gel. A Southern blot was performed with a probe specific for the minor satellite repeat (pMR150). The same blot was stripped and rehybridized with a mitochondrial DNA probe to demonstrate equal DNA loading. (F, right panels) Metaphase chromosomes from DCRΔ/Δ and Dicer-reconstituted cells were stained with a rabbit polyclonal antibody specific for trimethyl-H3K9. DAPI staining of chromosomes for each panel is shown on the left (100× magnification).
Similar articles
- The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb.
Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. Harfe BD, et al. Proc Natl Acad Sci U S A. 2005 Aug 2;102(31):10898-903. doi: 10.1073/pnas.0504834102. Epub 2005 Jul 22. Proc Natl Acad Sci U S A. 2005. PMID: 16040801 Free PMC article. - Dicer is essential for formation of the heterochromatin structure in vertebrate cells.
Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Fukagawa T, et al. Nat Cell Biol. 2004 Aug;6(8):784-91. doi: 10.1038/ncb1155. Epub 2004 Jul 11. Nat Cell Biol. 2004. PMID: 15247924 - Dicer is essential for mouse development.
Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Bernstein E, et al. Nat Genet. 2003 Nov;35(3):215-7. doi: 10.1038/ng1253. Epub 2003 Oct 5. Nat Genet. 2003. PMID: 14528307 - [Basic research on and application of RNA interference].
Ohta A, Inoue A, Taira K. Ohta A, et al. Gan To Kagaku Ryoho. 2004 Jun;31(6):827-31. Gan To Kagaku Ryoho. 2004. PMID: 15222096 Review. Japanese. - Gene silencing in human embryonic stem cells by RNA interference.
Rassouli FB, Matin MM. Rassouli FB, et al. Biochem Biophys Res Commun. 2009 Dec 25;390(4):1106-10. doi: 10.1016/j.bbrc.2009.10.038. Epub 2009 Oct 13. Biochem Biophys Res Commun. 2009. PMID: 19833094 Review.
Cited by
- Strategies for enrichment and selection of stem cell-derived tissue precursors.
Bernstein HS, Hyun WC. Bernstein HS, et al. Stem Cell Res Ther. 2012 May 10;3(3):17. doi: 10.1186/scrt108. Stem Cell Res Ther. 2012. PMID: 22575029 Free PMC article. Review. - Neurobehavioral Alterations in a Genetic Murine Model of Feingold Syndrome 2.
Fiori E, Babicola L, Andolina D, Coassin A, Pascucci T, Patella L, Han YC, Ventura A, Ventura R. Fiori E, et al. Behav Genet. 2015 Sep;45(5):547-59. doi: 10.1007/s10519-015-9724-8. Epub 2015 May 31. Behav Genet. 2015. PMID: 26026879 Free PMC article. - Human liver stem cell-derived microvesicles inhibit hepatoma growth in SCID mice by delivering antitumor microRNAs.
Fonsato V, Collino F, Herrera MB, Cavallari C, Deregibus MC, Cisterna B, Bruno S, Romagnoli R, Salizzoni M, Tetta C, Camussi G. Fonsato V, et al. Stem Cells. 2012 Sep;30(9):1985-98. doi: 10.1002/stem.1161. Stem Cells. 2012. PMID: 22736596 Free PMC article. - MicroRNA biomarkers for early detection of embryonic malformations in pregnancy.
Li X, Zhao Z. Li X, et al. J Biomol Res Ther. 2014 Dec;3(3):119. doi: 10.4172/2167-7956.1000119. J Biomol Res Ther. 2014. PMID: 25859419 Free PMC article. - The Role of Human Centromeric RNA in Chromosome Stability.
Leclerc S, Kitagawa K. Leclerc S, et al. Front Mol Biosci. 2021 Mar 31;8:642732. doi: 10.3389/fmolb.2021.642732. eCollection 2021. Front Mol Biosci. 2021. PMID: 33869284 Free PMC article. Review.
References
- Aravin A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11: 1017–1027. - PubMed
- Aravin A.A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5: 337–350. - PubMed
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
- Bernstein E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn, H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., and Hannon, G.J. 2003. Dicer is essential for mouse development. Nat. Genet. 35: 215–217. - PubMed
- Burdon T., Smith, A., and Savatier, P. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell Biol. 12: 432–438. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials