A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array - PubMed (original) (raw)
A sequence-based identification of the genes detected by probesets on the Affymetrix U133 plus 2.0 array
Jeremy Harbig et al. Nucleic Acids Res. 2005.
Abstract
One of the biggest problems facing microarray experiments is the difficulty of translating results into other microarray formats or comparing microarray results to other biochemical methods. We believe that this is largely the result of poor gene identification. We re-identified the probesets on the Affymetrix U133 plus 2.0 GeneChip array. This identification was based on the sequence of the probes and the sequence of the human genome. Using the BLAST program, we matched probes with documented and postulated human transcripts. This resulted in the redefinition of approximately 37% of the probes on the U133 plus 2.0 array. This updated identification specifically points out where the identification is complicated by cross-hybridization from splice variants or closely related genes. More than 5000 probesets detect multiple transcripts and therefore the exact protein affected cannot be readily concluded from the performance of one probeset alone. This makes naming difficult and impacts any downstream analysis such as associating gene ontologies, mapping affected pathways or simply validating expression changes. We have now automated the sequence-based identification and can more appropriately annotate any array where the sequence on each spot is known.
Figures
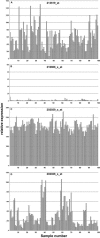
Figure 1
The signal captured by some probesets on the U133A array from 100 RNA samples collected from various tissues. Probeset 202029_x_at detects the expression of ribosomal protein L38. The other three probesets were designed to the complementary strand of the intended reference gene. Probeset 202028_s_at detects sequences complementary to the ribosomal protein L38. The plots for probesets 213619_at and 216868_s_at illustrate the difference between a probeset that detects a transcript and a probeset that does not detect a transcript. Although each plot is represented against a different scale, the relative expression levels are directly comparable.
Similar articles
- Quality assessment of the Affymetrix U133A&B probesets by target sequence mapping and expression data analysis.
Orlov YL, Zhou J, Lipovich L, Shahab A, Kuznetsov VA. Orlov YL, et al. In Silico Biol. 2007;7(3):241-60. In Silico Biol. 2007. PMID: 18415975 - Transcript-level annotation of Affymetrix probesets improves the interpretation of gene expression data.
Yu H, Wang F, Tu K, Xie L, Li YY, Li YX. Yu H, et al. BMC Bioinformatics. 2007 Jun 11;8:194. doi: 10.1186/1471-2105-8-194. BMC Bioinformatics. 2007. PMID: 17559689 Free PMC article. - Gene expression and isoform variation analysis using Affymetrix Exon Arrays.
Bemmo A, Benovoy D, Kwan T, Gaffney DJ, Jensen RV, Majewski J. Bemmo A, et al. BMC Genomics. 2008 Nov 7;9:529. doi: 10.1186/1471-2164-9-529. BMC Genomics. 2008. PMID: 18990248 Free PMC article. - Expression Profiling Using Affymetrix GeneChip Microarrays.
Auer H, Newsom DL, Kornacker K. Auer H, et al. Methods Mol Biol. 2009;509:35-46. doi: 10.1007/978-1-59745-372-1_3. Methods Mol Biol. 2009. PMID: 19212713 Review. - A comparison of analog and Next-Generation transcriptomic tools for mammalian studies.
Roy NC, Altermann E, Park ZA, McNabb WC. Roy NC, et al. Brief Funct Genomics. 2011 May;10(3):135-50. doi: 10.1093/bfgp/elr005. Epub 2011 Mar 9. Brief Funct Genomics. 2011. PMID: 21389008 Review.
Cited by
- A meta-analysis of kidney microarray datasets: investigation of cytokine gene detection and correlation with rt-PCR and detection thresholds.
Park WD, Stegall MD. Park WD, et al. BMC Genomics. 2007 Mar 30;8:88. doi: 10.1186/1471-2164-8-88. BMC Genomics. 2007. PMID: 17397532 Free PMC article. - Identification of molecular markers associated with the progression and prognosis of endometrial cancer: a bioinformatic study.
Liu J, Feng M, Li S, Nie S, Wang H, Wu S, Qiu J, Zhang J, Cheng W. Liu J, et al. Cancer Cell Int. 2020 Feb 19;20:59. doi: 10.1186/s12935-020-1140-3. eCollection 2020. Cancer Cell Int. 2020. PMID: 32099532 Free PMC article. - Misspellings or "miscellings"-Non-verifiable and unknown cell lines in cancer research publications.
Oste DJ, Pathmendra P, Richardson RAK, Johnson G, Ao Y, Arya MD, Enochs NR, Hussein M, Kang J, Lee A, Danon JJ, Cabanac G, Labbé C, Davis AC, Stoeger T, Byrne JA. Oste DJ, et al. Int J Cancer. 2024 Oct 1;155(7):1278-1289. doi: 10.1002/ijc.34995. Epub 2024 May 15. Int J Cancer. 2024. PMID: 38751110 - Global gene expression profiling of individual human oocytes and embryos demonstrates heterogeneity in early development.
Shaw L, Sneddon SF, Zeef L, Kimber SJ, Brison DR. Shaw L, et al. PLoS One. 2013 May 22;8(5):e64192. doi: 10.1371/journal.pone.0064192. Print 2013. PLoS One. 2013. PMID: 23717564 Free PMC article. - AffyMAPSDetector: a software tool to characterize Affymetrix GeneChip expression arrays with respect to SNPs.
Kumari S, Verma LK, Weller JW. Kumari S, et al. BMC Bioinformatics. 2007 Jul 30;8:276. doi: 10.1186/1471-2105-8-276. BMC Bioinformatics. 2007. PMID: 17663786 Free PMC article.
References
- Chuaqui R.F., Bonner R.F., Best C.J., Gillespie J.W., Flaig M.J., Hewitt S.M., Phillips J.L., Krizman D.B., Tangrea M.A., Ahram M., et al. Post-analysis follow-up and validation of microarray experiments. Nature Genet. 2002;32(Suppl.):509–514. - PubMed
- Ohyama H., Zhang X., Kohno Y., Alevizos I., Posner M., Wong D.T., Todd R. Laser capture microdissection-generated target sample for high-density oligonucleotide array hybridization. Biotechniques. 2000;29:530–536. - PubMed
- Iscove N.N., Barbara M., Gu M., Gibson M., Modi C., Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat. Biotechnol. 2002;20:940–943. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials