Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene - PubMed (original) (raw)
Transcription factor interactions and chromatin modifications associated with p53-mediated, developmental repression of the alpha-fetoprotein gene
Thi T Nguyen et al. Mol Cell Biol. 2005 Mar.
Abstract
We performed chromatin immunoprecipitation (ChIP) analyses of developmentally staged solid tissues isolated from wild-type and p53-null mice to determine specific histone N-terminal modifications, histone-modifying proteins, and transcription factor interactions at the developmental repressor region (-850) and core promoter of the hepatic tumor marker alpha-fetoprotein (AFP) gene. Both repression of AFP during liver development and silencing in the brain, where AFP is never expressed, are associated with dimethylation of histone H3 lysine 9 (DiMetH3K9) and the presence of heterochromatin protein 1 (HP1). These heterochromatic markers remain localized to AFP during developmental repression but spread to the upstream albumin gene during silencing. Developmentally regulated decreases in levels of acetylated H3 (AcH3K9) and H4 (AcH4) and of di- and trimethylated H3K4 (DiMetH3K4 and TriMetH3K4) occur at both the core promoter and distal repressor regions of AFP. Hepatic expression of AFP correlates with FoxA interaction at the repressor region and the binding of RNA polymerase II and TATA-binding protein to the core promoter. p53 acts as a developmental repressor of AFP in the liver by binding to chromatin, excluding FoxA interaction and targeting mSin3A/HDAC1 to the distal repressor region. p53-null mice exhibit developmentally delayed AFP repression, concomitant with acetylation of H3K9, methylation of H3K4, and loss of DiMetH3K9, mSin3A/HDAC1, and HP1 interactions.
Figures
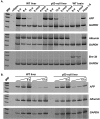
FIG. 1.
Tissue- and developmental-stage-specific regulation of AFP expression in WT and p53-null liver. (A) RT-PCR analysis was performed with RNAs isolated from liver tissues excised from 2-, 8-, and 12-day-old and 2- and 4-month-old WT and p53-null mice. Brain tissue RNAs from 8-day- and 2-month-old mice served as tissue-specific negative controls. Tumor-derived Hepa 1-6 cellular RNA served as a positive control for AFP expression. Primers specific for AFP, ALB, Brn-3b, and GAPDH were used to determine the relative levels of stable RNA expressed from each gene. (B) Multiple dilutions of input cDNA were analyzed as for panel A, over the linear range of PCR amplification. Levels of AFP expression were determined relative to GAPDH levels in the appropriate samples.
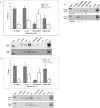
FIG. 2.
Changes in histone modifications during developmental repression of AFP conform to the paradigms of active and repressed chromatin. (A) ChIP analysis of the AFP developmental repressor region. (Top) Graph summarizing the results of multiple ChIP determinations. Each bar represents the average result from three to eight ChIP experiments; error bars, standard deviations. Cross-linked lysates were prepared from 8-day pp (gray bars) and 2-month (white bars) WT mouse liver tissue. Primers specific for the −850 SBE/p53RE were used in PCR amplification. Percent bound (% bound) represents a quantified value for antibody-precipitated, specific DNA divided by the amount of specific DNA present in the input lysate. (Bottom) Representative PCR analysis of the DNA present in chromatin fragments precipitated by each specific antibody, as indicated above each lane. Negative controls include nonspecific IgG, no antibody addition to the immunoprecipitation (no Ab), and no precipitated DNA added to the PCR (no DNA). An unmodified anti-histone H3 antibody (H3) served as a positive control. (B) ChIP analysis of the AFP start site region. (Top) Graph of multiple ChIP assays, as described for panel A. Primers specific for the AFP start site region were used in PCR amplification. (Bottom) Representative PCR analysis, as described for panel A. (C) PCR analysis of DiMetH3K4 and TriMetH3K4, in comparison to DiMetH3K9, associated with chromatin present at the AFP start site region in 8-day and 2-month WT mouse liver tissue.
FIG. 3.
Analysis of histone modifications present in permanently silenced and constitutively expressed chromatin isolated from brain and liver tissues. (A) ChIP analysis of silenced AFP chromatin at the SBE/p53RE and expressed Brn-3b chromatin isolated from brain tissue. (Top) Graph summarizing the results of multiple ChIP determinations, as described for Fig. 2A. Histone modifications associated with AFP chromatin (gray bars) and Brn-3b chromatin (white bars) in the brain were determined as described in Materials and Methods. (Bottom) Representative PCR analysis, as described for Fig. 2. Cross-linked chromatin lysates from 2-month WT mouse brain tissue were prepared as described. (B) ChIP determinations of histone modifications present in permanently silenced Brn-3b chromatin and constitutively activated ALB chromatin isolated from liver tissue at 8 days and 2 months of development. (Top) Graph summarizing the results of multiple ChIP determinations. Histone modifications were associated with specific genes as indicated. Each gene set is presented in the following order: ALB, 8-day (gray bar) and 2-month (white bar) liver; Brn-3b, 8-day (horizontal gradient bar) and 2-month (vertical gradient bar) liver. (Bottom) Representative PCR analysis. Cross-linked chromatin lysates from 2-month WT mouse liver tissue were prepared as described.
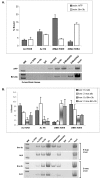
FIG. 4.
Inverse relationship between activators and repressors of transcription during developmental regulation of AFP. (A) ChIP analyses of developmentally staged liver tissue from 8-day-old and 2-month-old mice. Representative PCR analyses of chromatin fragments associated with the transactivator protein FoxA, RNA polymerase II [Pol II (ser5)], and the corepressor protein mSin3A. The presence of bound ALB enhancer, AFP SBE/p53RE, and the AFP start site region was determined and quantified relative to input (% bound). (B) TBP binding in relationship to AcH4 at the AFP transcription start site region during liver development, determined by ChIP.
FIG. 5.
HP1 associates with the AFP repressor and promoter regions during developmental repression and tissue-specific silencing, but HDAC1 binding is restricted to the developmental repressor. (A to C) Representative PCR analyses of ChIP studies for the association of HDAC1 and HP1 with the developmental repressor (SBE/p53RE) and start site regions of AFP in comparison to Brn-3b. Results of assays of 8-day (A) and 2-month (B) liver tissue and of 2-month brain tissue(C) are shown. Unmodified anti-histone H3 antibody is shown as an internal control for chromatin recovery in each lysate. Binding is quantified (% bound) in comparison to each input lysate; titrations were analyzed separately (data not shown). (D) Bar graph summarizing the developmental changes in H3K9 modifications and HP1 interactions during liver development and in brain tissue at specific regions of the ALB/AFP gene locus: the AFP start site, centered at +1; the SBE/p53RE, centered at −850; AFP enhancer (enh) III, at −7 kb; and the enhancer of ALB, at −11 kb relative to the ALB start site of transcription and approximately 27 kb upstream of AFP enhancer III. Data derived from ChIP analyses for which results are shown here and in Fig. 2 (also data not shown) were compared by calculation of the ratio between average bound protein and average chromatin recovery (percent unmodified histone H3) for each lysate. Averages of these ratios are shown as bars in the bar graph: light gray and dark gray, 8-day and 2-month AcH3K9 levels; light gray gradient and dark gray gradient, 8-day and 2-month diMetH3K9 levels; faceted gray and solid white, 8-day and 2-month HP1 levels; stippled gray, 2-month brain HP1 levels. Data from 2-month liver tissue only are provided for AFP enhancer III.
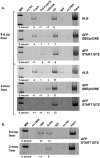
FIG. 6.
p53-dependent targeting of repression-associated modifications of histones and mSin3A/HDAC1 to AFP chromatin. (A) In the absence of p53, developmental repression of AFP is delayed. RNAs were isolated from 2- and 12-day pp and 1-, 2-, and 4-month livers extracted from WT and p53-null mice. RT-PCR was performed to determine relative levels of AFP and GAPDH expression. PCR amplification at 24 cycles was used to assay GAPDH levels (upper panel, GAPDH), and 30 cycles were used with the same RNA samples to determine AFP (lower panel, AFP). (B) Representative PCR determinations of ChIP assays of specific histone modifications and p53/mSin3A present at the SBE/p53RE site in 2-month WT and p53-null liver tissue. The amount of DNA associated with each specific protein is quantified (% bound) in comparison to each input lysate; titrations were analyzed separately. (C) ChIP assays of FoxA and HDAC1 association with the SBE/p53RE and start site regions of AFP in p53-null liver tissue. Representative data are shown. (D) Bar graph summarizing the p53-dependent patterns of FoxA, p53, mSin3A, and HDAC1 proteins bound to SBE/p53RE during developmental repression of AFP. Analyses of 8-day WT (light gray bars), 2-month WT (dark gray bars), and 2-month p53-null (white bars) livers were performed two to four times each. Data from these analyses are presented here and in Fig. 4 and 5 (also data not shown). Ratios of bound proteins to unmodified histone H3 were calculated for each lysate and averaged, as described for Fig. 5D.
Similar articles
- A direct intersection between p53 and transforming growth factor beta pathways targets chromatin modification and transcription repression of the alpha-fetoprotein gene.
Wilkinson DS, Ogden SK, Stratton SA, Piechan JL, Nguyen TT, Smulian GA, Barton MC. Wilkinson DS, et al. Mol Cell Biol. 2005 Feb;25(3):1200-12. doi: 10.1128/MCB.25.3.1200-1212.2005. Mol Cell Biol. 2005. PMID: 15657445 Free PMC article. - Family members p53 and p73 act together in chromatin modification and direct repression of alpha-fetoprotein transcription.
Cui R, Nguyen TT, Taube JH, Stratton SA, Feuerman MH, Barton MC. Cui R, et al. J Biol Chem. 2005 Nov 25;280(47):39152-60. doi: 10.1074/jbc.M504655200. Epub 2005 Oct 2. J Biol Chem. 2005. PMID: 16203738 - p53 targets chromatin structure alteration to repress alpha-fetoprotein gene expression.
Ogden SK, Lee KC, Wernke-Dollries K, Stratton SA, Aronow B, Barton MC. Ogden SK, et al. J Biol Chem. 2001 Nov 9;276(45):42057-62. doi: 10.1074/jbc.C100381200. Epub 2001 Sep 25. J Biol Chem. 2001. PMID: 11572852 - Maintaining memory of silencing at imprinted differentially methylated regions.
Voon HP, Gibbons RJ. Voon HP, et al. Cell Mol Life Sci. 2016 May;73(9):1871-9. doi: 10.1007/s00018-016-2157-6. Epub 2016 Feb 16. Cell Mol Life Sci. 2016. PMID: 26883803 Free PMC article. Review. - Histone deacetylases: silencers for hire.
Ng HH, Bird A. Ng HH, et al. Trends Biochem Sci. 2000 Mar;25(3):121-6. doi: 10.1016/s0968-0004(00)01551-6. Trends Biochem Sci. 2000. PMID: 10694882 Review.
Cited by
- Functional analysis of p53 binding under differential stresses.
Krieg AJ, Hammond EM, Giaccia AJ. Krieg AJ, et al. Mol Cell Biol. 2006 Oct;26(19):7030-45. doi: 10.1128/MCB.00322-06. Mol Cell Biol. 2006. PMID: 16980608 Free PMC article. - Chromatin-bound p53 anchors activated Smads and the mSin3A corepressor to confer transforming-growth-factor-beta-mediated transcription repression.
Wilkinson DS, Tsai WW, Schumacher MA, Barton MC. Wilkinson DS, et al. Mol Cell Biol. 2008 Mar;28(6):1988-98. doi: 10.1128/MCB.01442-07. Epub 2008 Jan 22. Mol Cell Biol. 2008. PMID: 18212064 Free PMC article. - Gadd45β is an inducible coactivator of transcription that facilitates rapid liver growth in mice.
Tian J, Huang H, Hoffman B, Liebermann DA, Ledda-Columbano GM, Columbano A, Locker J. Tian J, et al. J Clin Invest. 2011 Nov;121(11):4491-502. doi: 10.1172/JCI38760. Epub 2011 Oct 3. J Clin Invest. 2011. PMID: 21965327 Free PMC article. - Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells.
Taube JH, Allton K, Duncan SA, Shen L, Barton MC. Taube JH, et al. J Biol Chem. 2010 May 21;285(21):16135-44. doi: 10.1074/jbc.M109.088096. Epub 2010 Mar 26. J Biol Chem. 2010. PMID: 20348100 Free PMC article. - Widespread, exceptionally high levels of histone H3 lysine 4 trimethylation largely mediate "privileged" gene expression.
Chen L, Firozi P, Barton M, Templeton NS. Chen L, et al. Gene Expr. 2007;13(4-5):271-82. doi: 10.3727/000000006780666966. Gene Expr. 2007. PMID: 17605300 Free PMC article.
References
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. - PubMed
- Camper, S. A., R. Godbout, and S. M. Tilghman. 1989. The developmental regulation of albumin and alpha-fetoprotein gene expression. Prog. Nucleic Acid Res. Mol. Biol. 36:131-143. - PubMed
- Camper, S. A., and S. M. Tilghman. 1991. The activation and silencing of gene transcription in the liver. Bio/Technology 16:81-87. - PubMed
- Camper, S. A., and S. M. Tilghman. 1989. Postnatal repression of the alpha-fetoprotein gene is enhancer independent. Genes Dev. 3:537-546. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous