A computational study of off-target effects of RNA interference - PubMed (original) (raw)
A computational study of off-target effects of RNA interference
Shibin Qiu et al. Nucleic Acids Res. 2005.
Abstract
RNA interference (RNAi) is an intracellular mechanism for post-transcriptional gene silencing that is frequently used to study gene function. RNAi is initiated by short interfering RNA (siRNA) of approximately 21 nt in length, either generated from the double-stranded RNA (dsRNA) by using the enzyme Dicer or introduced experimentally. Following association with an RNAi silencing complex, siRNA targets mRNA transcripts that have sequence identity for destruction. A phenotype resulting from this knockdown of expression may inform about the function of the targeted gene. However, 'off-target effects' compromise the specificity of RNAi if sequence identity between siRNA and random mRNA transcripts causes RNAi to knockdown expression of non-targeted genes. The complete off-target effects must be investigated systematically on each gene in a genome by adjusting a group of parameters, which is too expensive to conduct experimentally and motivates a study in silico. This computational study examined the potential for off-target effects of RNAi, employing the genome and transcriptome sequence data of Homo sapiens, Caenorhabditis elegans and Schizosaccharomyces pombe. The chance for RNAi off-target effects proved considerable, ranging from 5 to 80% for each of the organisms, when using as parameter the exact identity between any possible siRNA sequences (arbitrary length ranging from 17 to 28 nt) derived from a dsRNA (range 100-400 nt) representing the coding sequences of target genes and all other siRNAs within the genome. Remarkably, high-sequence specificity and low probability for off-target reactivity were optimally balanced for siRNA of 21 nt, the length observed mostly in vivo. The chance for off-target RNAi increased (although not always significantly) with greater length of the initial dsRNA sequence, inclusion into the analysis of available untranslated region sequences and allowing for mismatches between siRNA and target sequences. siRNA sequences from within 100 nt of the 5' termini of coding sequences had low chances for off-target reactivity. This may be owing to coding constraints for signal peptide-encoding regions of genes relative to regions that encode for mature proteins. Off-target distribution varied along the chromosomes of C.elegans, apparently owing to the use of more unique sequences in gene-dense regions. Finally, biological and thermodynamical descriptors of effective siRNA reduced the number of potential siRNAs compared with those identified by sequence identity alone, but off-target RNAi remained likely, with an off-target error rate of approximately 10%. These results also suggest a direction for future in vivo studies that could both help in calibrating true off-target rates in living organisms and also in contributing evidence toward the debate of whether siRNA efficacy is correlated with, or independent of, the target molecule. In summary, off-target effects present a real but not prohibitive concern that should be considered for RNAi experiments.
Figures
Figure 1
Graphic depiction of variables tested computationally to investigate chance of off-target effect in each of the three organisms (H.sapiens, C.elegans and S.pombe). (A) General considerations: a target sequence (representing one particular expressed mRNA) is used as the source of dsRNA of which a pool of all possible siRNA is derived (mimicking the action of Dicer). Each sequence within the siRNA pool was compared for sequence identity (exact: m = 0; with mismatch: m > 0) to all possible siRNA sequences in the transcriptome through the feature map Φ(·) to determine chance of off-target errors. The parameters tested are as follows: (B) length of siRNA (n); (C) length of dsRNA (l); (D) addition of available UTR data in the target sequences (_u_3 and _u_5); and (E) position of the dsRNA along the target sequence (pos).
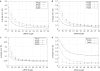
Figure 2
Effect of siRNA length and dsRNA length on off-target error rates under an exact match (m = 0) siRNA homology function. (A) H.sapiens and (B) C.elegans. (C) S.pombe, l = 100–400, n = 17–28. (D) Comparison of the three organisms for l = 300, n = 17–28 and m = 0. Within the length range tested, siRNA of 21 nt optimally combined target specificity and minimum length. Chance for off-target error increases significantly for smaller siRNAs, yet is not significantly different from longer siRNAs. Increased length of dsRNA yielded more diverse pools of siRNA, resulting in increased chance for off-target errors.
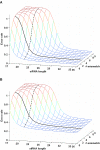
Figure 3
Effect of mismatch on RNAi off-target error rate in (A) C.elegans and (B) H.sapiens. Allowing for mismatches significantly increased the off-target error for siRNA of various lengths derived from CDSs of the transcriptome. Results shown were derived for l = 200, n = 17–35 and m = 0–9. Dashed curves indicate the positions of the planes of n = 21 and solid curves indicate the positions of the planes of m = 3 in the three-dimensional plots.
Figure 4
Effect of mismatch positions in H.sapiens, C.elegans and S.pombe. Mismatches in the region of 2–9 nt at the 5′ end of the guiding strand reduced off-target chances. Results shown were derived for n = 21, l = 100, m = 3 and mpos = 1–19.
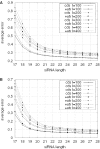
Figure 5
Effect of including UTR sequences with the CDS data for RNAi off-target error in (A) H.sapiens and (B) C.elegans. The off-target error for siRNA (various lengths) derived from CDSs of the transcriptome did not change significantly when available UTR sequence data were included among the sequences considered as the transcriptome (average _P_-value = 82%); l = 100–400, n = 17–28 and m = 0. CDS indicates the case where UTR is not included; ‘+UTR’ indicates the case where 3′-UTR is included.
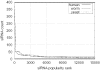
Figure 6
siRNA frequency distribution. The _x_-axis is the popularity rank of the siRNAs (the most frequently occurred siRNA is the most popular one and has the smallest abscissa). The _y_-axis is the corresponding count with which the siRNA occurs in the genome. The curves have very long and flat tails and the _x_-axis is actually only 1000th of the total data.
Figure 7
RNAi off-target error relative to varied position of the dsRNA along the target sequences. The off-target error is significantly lower when targeting the beginning 100–140 nt by dsRNA for generating siRNA. The _x_-axis is the start position of the dsRNA, pos. The _y_-axis is the error rate. (A) For C.elegans, off-target error shown for dsRNA lengths l = 100 and 200 nt, length of siRNA n = 22 and position pos 0–600. (B) Off-target error as a function of dsRNA position effect in H.sapiens (l = 100, 200, n = 25). (C) For S.pombe l = 200, 300, n = 25. When pos ≈ 100, the error reaches a local minimum.
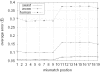
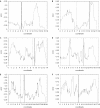
Figure 8
The distribution of RNAi off-target error of each gene plotted against the position of genes on the physical map of chromosomes of C.elegans (A–F represent chromosomes I–V and X, respectively). These plots disclosed that the chromosomal position is predictive of the off-target error rate. The curves were smoothed by averaging the off-target errors between neighboring genes. Dashed bars indicate physical center of the chromosomes (cM = 0), solid bars identify the position where most genes are centered (density center). Parameters are l = 300, n = 21, mpos = 0. Unit on the _x_-axis is million nt.
Figure 9
Effect of rational siRNA design in H.sapiens and C.elegans. (A) H.sapiens and (B) C.elegans. Results shown for l = 300, n = 17–29 (only odd number of lengths are considered since rational design rules need to compute the center of an siRNA), pool size = 5, 10, 20 and rational filter is also considered. The selection of siRNA with properties that were empirically found to be associated with highly functional siRNA sequences reduced off-target error relative to that of non-filtered siRNA. Note that off-target error is never reduced to zero.
Similar articles
- Novel insights into RNAi off-target effects using C. elegans paralogs.
Rual JF, Klitgord N, Achaz G. Rual JF, et al. BMC Genomics. 2007 Apr 19;8:106. doi: 10.1186/1471-2164-8-106. BMC Genomics. 2007. PMID: 17445269 Free PMC article. - RNA string kernels for RNAi off-target evaluation.
Qiu S, Lane T. Qiu S, et al. Int J Bioinform Res Appl. 2006;2(2):132-46. doi: 10.1504/IJBRA.2006.009764. Int J Bioinform Res Appl. 2006. PMID: 18048158 - siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect.
Naito Y, Yoshimura J, Morishita S, Ui-Tei K. Naito Y, et al. BMC Bioinformatics. 2009 Nov 30;10:392. doi: 10.1186/1471-2105-10-392. BMC Bioinformatics. 2009. PMID: 19948054 Free PMC article. - Structural diversity repertoire of gene silencing small interfering RNAs.
Chang CI, Kim HA, Dua P, Kim S, Li CJ, Lee DK. Chang CI, et al. Nucleic Acid Ther. 2011 Jun;21(3):125-31. doi: 10.1089/nat.2011.0286. Nucleic Acid Ther. 2011. PMID: 21749289 Free PMC article. Review. - RNAi mechanisms in Caenorhabditis elegans.
Grishok A. Grishok A. FEBS Lett. 2005 Oct 31;579(26):5932-9. doi: 10.1016/j.febslet.2005.08.001. Epub 2005 Aug 9. FEBS Lett. 2005. PMID: 16162338 Review.
Cited by
- First knockdown gene expression in bat (Hipposideros armiger) brain mediated by lentivirus.
Chen Q, Zhu T, Jones G, Zhang J, Sun Y. Chen Q, et al. Mol Biotechnol. 2013 Jun;54(2):564-71. doi: 10.1007/s12033-012-9596-6. Mol Biotechnol. 2013. PMID: 22965420 - Crowdsourced Identification of Potential Target Genes for CTV Induced Gene Silencing for Controlling the Citrus Greening Vector Diaphorina citri.
Ramos JE, Jain RG, Powell CA, Dawson WO, Gowda S, Borovsky D, Shatters RG Jr. Ramos JE, et al. Front Physiol. 2021 Apr 9;12:571826. doi: 10.3389/fphys.2021.571826. eCollection 2021. Front Physiol. 2021. PMID: 33897443 Free PMC article. - RNAi-Based Therapy: Combating Shrimp Viral Diseases.
Alam MS, Islam MN, Das M, Islam SF, Rabbane MG, Karim E, Roy A, Alam MS, Ahmed R, Kibria ASM. Alam MS, et al. Viruses. 2023 Oct 5;15(10):2050. doi: 10.3390/v15102050. Viruses. 2023. PMID: 37896827 Free PMC article. Review. - Antisense oligodeoxynucleotide inhibition as an alternative and convenient method for gene function analysis in pollen tubes.
Liao F, Wang L, Yang LB, Zhang L, Peng X, Sun MX. Liao F, et al. PLoS One. 2013;8(3):e59112. doi: 10.1371/journal.pone.0059112. Epub 2013 Mar 20. PLoS One. 2013. PMID: 23527102 Free PMC article. - In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference.
Jiang Y, Loker ES, Zhang SM. Jiang Y, et al. Dev Comp Immunol. 2006;30(10):855-66. doi: 10.1016/j.dci.2005.12.004. Epub 2006 Jan 10. Dev Comp Immunol. 2006. PMID: 16442620 Free PMC article.
References
- Fire A., Xu S.Q., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
- Fraser A.J.G., Kamath R.S., Zipperten P., Campos M.M., Sohrmann M., Ahringer J. Functional genomic analysis of C.elegans chromosome I by systemic RNA interference. Nature. 2000;408:325–330. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials