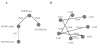Filtering high-throughput protein-protein interaction data using a combination of genomic features - PubMed (original) (raw)
Filtering high-throughput protein-protein interaction data using a combination of genomic features
Ashwini Patil et al. BMC Bioinformatics. 2005.
Abstract
Background: Protein-protein interaction data used in the creation or prediction of molecular networks is usually obtained from large scale or high-throughput experiments. This experimental data is liable to contain a large number of spurious interactions. Hence, there is a need to validate the interactions and filter out the incorrect data before using them in prediction studies.
Results: In this study, we use a combination of 3 genomic features -- structurally known interacting Pfam domains, Gene Ontology annotations and sequence homology -- as a means to assign reliability to the protein-protein interactions in Saccharomyces cerevisiae determined by high-throughput experiments. Using Bayesian network approaches, we show that protein-protein interactions from high-throughput data supported by one or more genomic features have a higher likelihood ratio and hence are more likely to be real interactions. Our method has a high sensitivity (90%) and good specificity (63%). We show that 56% of the interactions from high-throughput experiments in Saccharomyces cerevisiae have high reliability. We use the method to estimate the number of true interactions in the high-throughput protein-protein interaction data sets in Caenorhabditis elegans, Drosophila melanogaster and Homo sapiens to be 27%, 18% and 68% respectively. Our results are available for searching and downloading at http://helix.protein.osaka-u.ac.jp/htp/.
Conclusion: A combination of genomic features that include sequence, structure and annotation information is a good predictor of true interactions in large and noisy high-throughput data sets. The method has a very high sensitivity and good specificity and can be used to assign a likelihood ratio, corresponding to the reliability, to each interaction.
Figures
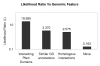
Figure 1
Likelihood ratios for genomic features.
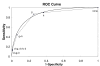
Figure 2
ROC curve for the combination of genomic features using 10-fold cross validations. The dotted line shows the empirical ROC curve, while the solid line shows the fitted ROC curve (obtained using JROCFIT). Each point on the ROC curve corresponds to sensitivity and specificity for one or a combination of more than one genomic features. d: interacting Pfam domains; g: similar GO annotations; h: homologous interactions; none: no genomic features. More than one genomic features are indicated by listing the features separated by a '+' sign.
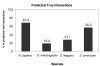
Figure 3
Percentage of interactions predicted true across different high-throughput data sets.
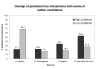
Figure 4
Percentage of interactions predicted true in high and low confidence interactions across different high-throughput data sets.
Figure 5
Some low confidence interactions predicted to be true by our method and confirmed by other publications. The Likelihood ratio for each interaction is indicated. Interactions with a Likelihood ratio greater than 100 are shown with a solid line, while those with a Likelihood ratio less than 10 are shown with a dashed line. (A) Interactions between proteins co-regulating the alternative splicing of Dscam exon 4 in D. menalogaster. (B) Interactions between proteins in the Lsm1-7 complex in S. cerevisiae confirmed by similar interactions found in H. sapiens.
Similar articles
- Genome-scale gene function prediction using multiple sources of high-throughput data in yeast Saccharomyces cerevisiae.
Joshi T, Chen Y, Becker JM, Alexandrov N, Xu D. Joshi T, et al. OMICS. 2004 Winter;8(4):322-33. doi: 10.1089/omi.2004.8.322. OMICS. 2004. PMID: 15703479 - AVID: an integrative framework for discovering functional relationships among proteins.
Jiang T, Keating AE. Jiang T, et al. BMC Bioinformatics. 2005 Jun 1;6:136. doi: 10.1186/1471-2105-6-136. BMC Bioinformatics. 2005. PMID: 15929793 Free PMC article. - VisANT: an online visualization and analysis tool for biological interaction data.
Hu Z, Mellor J, Wu J, DeLisi C. Hu Z, et al. BMC Bioinformatics. 2004 Feb 19;5:17. doi: 10.1186/1471-2105-5-17. BMC Bioinformatics. 2004. PMID: 15028117 Free PMC article. - Conservation of protein-protein interactions - lessons from ascomycota.
Pagel P, Mewes HW, Frishman D. Pagel P, et al. Trends Genet. 2004 Feb;20(2):72-6. doi: 10.1016/j.tig.2003.12.007. Trends Genet. 2004. PMID: 14746987 Review. - The Cartographers toolbox: building bigger and better human protein interaction networks.
Sanderson CM. Sanderson CM. Brief Funct Genomic Proteomic. 2009 Jan;8(1):1-11. doi: 10.1093/bfgp/elp003. Epub 2009 Mar 12. Brief Funct Genomic Proteomic. 2009. PMID: 19282470 Review.
Cited by
- Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system.
Stynen B, Tournu H, Tavernier J, Van Dijck P. Stynen B, et al. Microbiol Mol Biol Rev. 2012 Jun;76(2):331-82. doi: 10.1128/MMBR.05021-11. Microbiol Mol Biol Rev. 2012. PMID: 22688816 Free PMC article. Review. - Reconstruction of protein-protein interaction network of insulin signaling in Homo sapiens.
Durmuş Tekir S, Ümit P, Eren Toku A, Ülgen KÖ. Durmuş Tekir S, et al. J Biomed Biotechnol. 2010;2010:690925. doi: 10.1155/2010/690925. Epub 2010 Dec 14. J Biomed Biotechnol. 2010. PMID: 21197403 Free PMC article. - Mapping the Synthetic Dosage Lethality Network of CDK1/CDC28.
Zimmermann C, Garcia I, Omerzu M, Chymkowitch P, Zhang B, Enserink JM. Zimmermann C, et al. G3 (Bethesda). 2017 Jun 7;7(6):1753-1766. doi: 10.1534/g3.117.042317. G3 (Bethesda). 2017. PMID: 28428242 Free PMC article. - Triangle network motifs predict complexes by complementing high-error interactomes with structural information.
Andreopoulos B, Winter C, Labudde D, Schroeder M. Andreopoulos B, et al. BMC Bioinformatics. 2009 Jun 27;10:196. doi: 10.1186/1471-2105-10-196. BMC Bioinformatics. 2009. PMID: 19558694 Free PMC article. - PPISearch: a web server for searching homologous protein-protein interactions across multiple species.
Chen CC, Lin CY, Lo YS, Yang JM. Chen CC, et al. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W369-75. doi: 10.1093/nar/gkp309. Epub 2009 May 5. Nucleic Acids Res. 2009. PMID: 19417070 Free PMC article.
References
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. - DOI - PubMed
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases