UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus - PubMed (original) (raw)
Comparative Study
. 2005 Jun 14;102(24):8650-5.
doi: 10.1073/pnas.0501458102. Epub 2005 Jun 6.
Affiliations
- PMID: 15939881
- PMCID: PMC1150825
- DOI: 10.1073/pnas.0501458102
Comparative Study
UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus
Yachuan Yu et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Chromatin immunoprecipitation with anti-acetyl histone H3 (K9 and K14) and anti-acetyl histone H4 (K5, K8, K12, and K16) antibodies shows that Lys-9 and/or Lys-14 of histone H3, but not the relevant sites of histone H4 in nucleosomes at the repressed MFA2 promoter, are hyperacetylated after UV irradiation. This level of histone hyperacetylation diminishes gradually as repair proceeds. Accompanying this, chromatin in the promoter becomes more accessible to restriction enzymes after UV irradiation and returns to the pre-UV state gradually. UV-related histone hyperacetylation and chromatin remodeling in the MFA2 promoter depend on Gcn5p and partially on Swi2p, respectively. Deletion of GCN5, but not of SWI2, impairs repair of DNA damage in the MFA2 promoter. The post-UV histone modifications and chromatin remodeling at the repressed MFA2 promoter do not activate MFA2 transcriptionally, nor do they require damage recognition by Rad4p or Rad14p. Furthermore, we show that UV irradiation triggers genome-wide histone hyperacetylation at both histone H3 and H4. These experiments indicate that chromatin at a yeast repressed locus undergoes active change after UV radiation treatment and that failure to achieve histone H3 hyperacetylation impairs the repair of DNA damage.
Figures
Fig. 1.
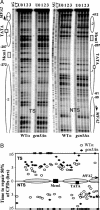
Repair of CPDs in the MFA2 promoter in α-mating type strains. (A) Gels depicting CPDs in the transcribed strand and nontranscribed strand of HaeIII restriction fragment (-516 to +83) in the repressed MFA2 promoter in wild-type (WT) and the _gcn5_Δ α strains after 150 J/m2 UV irradiation. Lane U, DNA from nonirradiated cells; lanes 0-3, DNA from irradiated cells after 0-3 h of repair. Alongside the gels are symbols representing nucleosome positions, MFA2 upstream activating sequences, and the start site of MFA2 transcription (arrow). Nucleotide positions are allocated in relation to the MFA2 start codon. (B) Time to remove 50% of the initial CPDs (_T_50%) at given sites. _T_50% of a single CPD or a group of CPDs with a similar repair rate was calculated (<3 h) or extrapolated (>3 h) as described in ref. . The _T_50% of slowly repaired CPDs (_T_50% ≥ 6 h) was shown at the same level (≥6 h) on the graph.
Fig. 2.
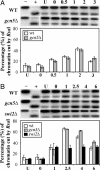
Post-UV histone acetylation. The level of acetylation is presented as the fold increase relative to that in unirradiated wild-type α cells. At the top A, B, and C are the acetylated sites. PCR was performed to amplify a region of 135 bp in nucleosome -2 and a region of 122 bp in nucleosome -1 in the MFA2 promoter. These two close regions gave the same result. (A and B) The acetylation of histones H3 and H4 in the repressed MFA2 promoter before and after 150 J/m2 UV irradiation. (C) The acetylation of histone H3 in the repressed MFA2 promoter of wild-type cells in response to 75 J/m2 UV irradiation. (D) Total histone acetylation in response to 150 J/m2 UV irradiation. (Top) Gels stained with Coomassie brilliant blue to indicate total histones. (Middle and Bottom) Western blots probed with either anti-acetyl histone H3 (K9 and K14; Upstate Biotechnology) or anti-acetyl histone H4 (K5, K8, K12, and K16; Upstate Biotechnology). Lane Ac+, positive control for the acetylated histones from sodium butyrate-treated cells (see Materials and Methods).
Fig. 3.
The accessibility of the RsaI site in chromatin in the repressed MFA2 promoter in response to 150 J/m2 (A) and 400 J/m2 (B) UV irradiation. Lane -, naked DNA digested by HaeIII only; lane +, naked DNA digested by both HaeIII and RsaI. The remainders are recovered DNAs after the sequential restriction enzyme digestion of chromatin (see Materials and Methods) from unirradiated (U) and UV-irradiated cells after various repair times in hours. Below the gels are the quantitative data presented as the percentage of chromatin being cut by RsaI.
Fig. 4.
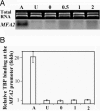
Transcription of MFA2 after UV radiation. (A) Northern blots of MFA2 mRNA. Lane A, RNA from a mating type cells where MFA2 is transcribed; lane U: RNA from unirradiated α cells; lanes 0, 0.5, 1, and 2, RNA from UV-irradiated (150 J/m2) cells after various repair times as indicated in hours. (B) The occupancy of TBP at the MFA2 promoter. Lane A (active), binding of TBP at the MFA2 promoter when MFA2 is active; lane U, occupancy of TBP at the repressed MFA2 promoter in α cells; lanes 0, 1, and 2, after UV following the repair times in hours.
Fig. 5.
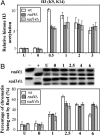
Histone H3 acetylation and accessibility of the RsaI site in chromatin at the repressed MFA2 promoter in the _rad4_Δ and _rad14_Δ α mutants. (A) Histone H3 acetylation (K9 and K14) in response to 150 J/m2 UV irradiation. The histone H3 acetylation level is presented as the fold increase relative to that before UV irradiation (U). The remaining samples are from UV-treated cells after repair from 0-3 h. (B) Accessibility of DNA in chromatin to RsaI in response to 400 J/m2 UV irradiation. The accessibility of DNA in chromatin is represented by the percentage of chromatin sensitive to RsaI at the MFA2 promoter. Lane -, naked DNA digested by HaeIII only; lane +, naked DNA digested by both HaeIII and RsaI; lane U, chromatin sample with no UV; lanes 0-6, chromatin samples from cells receiving 400 J/m2 UV irradiation after various repair times in hours.
Similar articles
- Histone acetylation, chromatin remodelling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae.
Yu Y, Waters R. Yu Y, et al. Cell Cycle. 2005 Aug;4(8):1043-5. doi: 10.4161/cc.4.8.1928. Epub 2005 Aug 21. Cell Cycle. 2005. PMID: 16082210 Review. - Histone acetylation, chromatin remodelling, transcription and nucleotide excision repair in S. cerevisiae: studies with two model genes.
Teng Y, Yu Y, Ferreiro JA, Waters R. Teng Y, et al. DNA Repair (Amst). 2005 Jul 28;4(8):870-83. doi: 10.1016/j.dnarep.2005.04.006. DNA Repair (Amst). 2005. PMID: 15950549 - How chromatin is remodelled during DNA repair of UV-induced DNA damage in Saccharomyces cerevisiae.
Yu S, Teng Y, Waters R, Reed SH. Yu S, et al. PLoS Genet. 2011 Jun;7(6):e1002124. doi: 10.1371/journal.pgen.1002124. Epub 2011 Jun 16. PLoS Genet. 2011. PMID: 21698136 Free PMC article. - Open, repair and close again: chromatin dynamics and the response to UV-induced DNA damage.
Palomera-Sanchez Z, Zurita M. Palomera-Sanchez Z, et al. DNA Repair (Amst). 2011 Feb 7;10(2):119-25. doi: 10.1016/j.dnarep.2010.10.010. Epub 2010 Dec 3. DNA Repair (Amst). 2011. PMID: 21130713 Review.
Cited by
- Trichostatin A Promotes Cytotoxicity of Cisplatin, as Evidenced by Enhanced Apoptosis/Cell Death Markers.
Zhou Y, Luo Q, Zeng F, Liu X, Han J, Gu L, Tian X, Zhang Y, Zhao Y, Wang F. Zhou Y, et al. Molecules. 2024 Jun 3;29(11):2623. doi: 10.3390/molecules29112623. Molecules. 2024. PMID: 38893499 Free PMC article. - DNA Repair in Nucleosomes: Insights from Histone Modifications and Mutants.
Selvam K, Wyrick JJ, Parra MA. Selvam K, et al. Int J Mol Sci. 2024 Apr 16;25(8):4393. doi: 10.3390/ijms25084393. Int J Mol Sci. 2024. PMID: 38673978 Free PMC article. Review. - Catching Nucleosome by Its Decorated Tails Determines Its Functional States.
Sehrawat P, Shobhawat R, Kumar A. Sehrawat P, et al. Front Genet. 2022 Jul 14;13:903923. doi: 10.3389/fgene.2022.903923. eCollection 2022. Front Genet. 2022. PMID: 35910215 Free PMC article. Review. - Microbial Adaptation to Enhance Stress Tolerance.
Tan YS, Zhang RK, Liu ZH, Li BZ, Yuan YJ. Tan YS, et al. Front Microbiol. 2022 Apr 27;13:888746. doi: 10.3389/fmicb.2022.888746. eCollection 2022. Front Microbiol. 2022. PMID: 35572687 Free PMC article. Review. - Epigenetic Regulation of Nucleotide Excision Repair.
Li W, Jones K, Burke TJ, Hossain MA, Lariscy L. Li W, et al. Front Cell Dev Biol. 2022 Apr 8;10:847051. doi: 10.3389/fcell.2022.847051. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35465333 Free PMC article. Review.
References
- Peterson, C. L. & Workman, J. L. (2000) Curr. Opin. Genet. Dev. 10, 187-192. - PubMed
- Berger, S. L. (2002) Curr. Opin. Genet. Dev. 12, 142-148. - PubMed
- de Laat, W. L., Jaspers, N. G. & Hoeijmakers, J. H. (1999) Genes Dev. 13, 768-785. - PubMed
- Reed, S. H. & Waters, R. (2003) in Nature Encyclopaedia of the Human Genome, ed. Cooper, D. N. (Nature Publishing Group, London), Vol. 2, pp. 148-154.
- Wang, Z.G., Wu, X. H. & Friedberg, E. C. (1991) J. Biol. Chem. 266, 22472-22478. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials