Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains - PubMed (original) (raw)
Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains
S Balaji et al. Nucleic Acids Res. 2005.
Abstract
The comparative genomics of apicomplexans, such as the malarial parasite Plasmodium, the cattle parasite Theileria and the emerging human parasite Cryptosporidium, have suggested an unexpected paucity of specific transcription factors (TFs) with DNA binding domains that are closely related to those found in the major families of TFs from other eukaryotes. This apparent lack of specific TFs is paradoxical, given that the apicomplexans show a complex developmental cycle in one or more hosts and a reproducible pattern of differential gene expression in course of this cycle. Using sensitive sequence profile searches, we show that the apicomplexans possess a lineage-specific expansion of a novel family of proteins with a version of the AP2 (Apetala2)-integrase DNA binding domain, which is present in numerous plant TFs. About 20-27 members of this apicomplexan AP2 (ApiAP2) family are encoded in different apicomplexan genomes, with each protein containing one to four copies of the AP2 DNA binding domain. Using gene expression data from Plasmodium falciparum, we show that guilds of ApiAP2 genes are expressed in different stages of intraerythrocytic development. By analogy to the plant AP2 proteins and based on the expression patterns, we predict that the ApiAP2 proteins are likely to function as previously unknown specific TFs in the apicomplexans and regulate the progression of their developmental cycle. In addition to the ApiAP2 family, we also identified two other novel families of AP2 DNA binding domains in bacteria and transposons. Using structure similarity searches, we also identified divergent versions of the AP2-integrase DNA binding domain fold in the DNA binding region of the PI-SceI homing endonuclease and the C-terminal domain of the pleckstrin homology (PH) domain-like modules of eukaryotes. Integrating these findings, we present a reconstruction of the evolutionary scenario of the AP2-integrase DNA binding domain fold, which suggests that it underwent multiple independent combinations with different types of mobile endonucleases or recombinases. It appears that the eukaryotic versions have emerged from versions of the domain associated with mobile elements, followed by independent lineage-specific expansions, which accompanied their recruitment to transcription regulation functions.
Figures
Figure 1
Alignment of AP2 domains. Proteins are denoted by their gene names, species abbreviations and GenBank identifier (gi) numbers. The number of AP2 domains in a polypeptide is shown to the right of the alignment. Residues involved in contacting DNA in the solution structure of the AP2 domain (pdb id: 1GCC) are shown below the alignment. The secondary structure was derived from the solution structure of the AP2 domain (PDB ID: 1GCC). E represents a β strand; H, helix. The coloring reflects the conservation profile at 80% consensus. The coloring scheme and consensus abbreviations are as follows: h, hydrophobic (h: ACFILMVWY) and a, aromatic (a: FWY) residues shaded yellow; b, big (LIYERFQKMW) residues shaded gray, s, small (AGSVCDN) residues colored green; and p, polar (STEDKRNQHC) residues colored magenta. Species abbreviations are as follows: APMV: Acanthamoeba polyphaga mimivirus; Atha: A.thaliana; Atum: Agrobacterium tumefaciens; BP01: Bacteriophage Felix 01; BPCorn: Mycobacteriophage Corndog; BPHK022: Enterobacteria phage HK022; BPRB49: Enterobacteria phage RB49; BPST3: Streptococcus thermophilus bacteriophage ST3; BPT1: Enterobacteria phage T1; BPT5: Bacteriophage T5; BPT7: Enterobacteria phage T7; BPXp10: X.oryzae bacteriophage Xp10; BPphig1e: Bacteriophage phig1e; Caur: Chloroflexus aurantiacus; Chom: Cryptosporidium hominis; Cpar: C.parvum; Dpsy: D.psychrophila; Ecol: Escherichia coli; Efae: Enterococcus faecalis; Ghir: Gossypium hirsutum; Lesc: Lycopersicon esculentum; Lmon: Listeria monocytogenes; Lpla: Lactobacillus plantarum; Nsyl: Nicotiana sylvestris; Pfa: Plasmodium falciparum; Rbal: Rhodopirellula baltica; Spyo: Streptococcus pyogenes; Taes: Triticum aestivum; Theileria annulata; Tery: Trichodesmium erythraeum; Tfus: Thermobifida fusca; Tthe: Tetrahymena thermophila; Vvul: Vibrio vulnificus.
Figure 1
Alignment of AP2 domains. Proteins are denoted by their gene names, species abbreviations and GenBank identifier (gi) numbers. The number of AP2 domains in a polypeptide is shown to the right of the alignment. Residues involved in contacting DNA in the solution structure of the AP2 domain (pdb id: 1GCC) are shown below the alignment. The secondary structure was derived from the solution structure of the AP2 domain (PDB ID: 1GCC). E represents a β strand; H, helix. The coloring reflects the conservation profile at 80% consensus. The coloring scheme and consensus abbreviations are as follows: h, hydrophobic (h: ACFILMVWY) and a, aromatic (a: FWY) residues shaded yellow; b, big (LIYERFQKMW) residues shaded gray, s, small (AGSVCDN) residues colored green; and p, polar (STEDKRNQHC) residues colored magenta. Species abbreviations are as follows: APMV: Acanthamoeba polyphaga mimivirus; Atha: A.thaliana; Atum: Agrobacterium tumefaciens; BP01: Bacteriophage Felix 01; BPCorn: Mycobacteriophage Corndog; BPHK022: Enterobacteria phage HK022; BPRB49: Enterobacteria phage RB49; BPST3: Streptococcus thermophilus bacteriophage ST3; BPT1: Enterobacteria phage T1; BPT5: Bacteriophage T5; BPT7: Enterobacteria phage T7; BPXp10: X.oryzae bacteriophage Xp10; BPphig1e: Bacteriophage phig1e; Caur: Chloroflexus aurantiacus; Chom: Cryptosporidium hominis; Cpar: C.parvum; Dpsy: D.psychrophila; Ecol: Escherichia coli; Efae: Enterococcus faecalis; Ghir: Gossypium hirsutum; Lesc: Lycopersicon esculentum; Lmon: Listeria monocytogenes; Lpla: Lactobacillus plantarum; Nsyl: Nicotiana sylvestris; Pfa: Plasmodium falciparum; Rbal: Rhodopirellula baltica; Spyo: Streptococcus pyogenes; Taes: Triticum aestivum; Theileria annulata; Tery: Trichodesmium erythraeum; Tfus: Thermobifida fusca; Tthe: Tetrahymena thermophila; Vvul: Vibrio vulnificus.
Figure 2
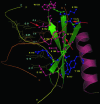
Structures of different domains of the AP2-IDBD fold. Strands and helices of the AP2-IDBD fold are colored green and pink, respectively. PDB ids for the displayed structures as follows; 1gcc: GCC-box binding domain; 1bb8: tn916 integrase DNA binding domain; 1kjk: lambda integrase N-terminal domain; 1qqg: Insulin receptor substrate 1 (IRS-1); 1lwt: PI-SceI homing endonuclease DNA binding domain.
Figure 3
DNA interactions of the AP2 domain. The solution structure of the A.thaliana GCC-box binding domain in complex with DNA (PDB Id: 1gcc) is shown. Strands are colored green and the helix is colored pink. Complementary DNA strands are labeled I and II and colored orange and yellow, respectively. The side-chains of DNA-contacting residues are displayed in the ball and stick format. Residues that interact with DNA bases are colored pink and those that predominantly interact with the DNA backbone are colored blue. Red arrows indicate positions that are well conserved in the ApiAP2 family (see Figure 1 and Table 1 for the equivalent residues in the ApiAP2 proteins).
Figure 4
Domain architectures of AP2 domain proteins. Domains are represented by their standard notations. ATH represents the AT-hook. The protein naming scheme and species abbreviations are as in Figure 1.
Figure 5
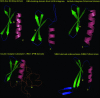
Expression patterns of AP2 proteins. Stage-specific expression of the ApiAp2 TFs and their potential target genes during the IDC. Microarray gene expression data were available for 46 timepoints as shown (26). Using _K_-means clustering, the predicted ApiAp2 TFs were grouped into five clusters. The first four clusters correspond to the four major developmental stages: (a) ring (b) trophozoite (c) early schizont and (d) schizont, whereas the fifth cluster (e) consists of genes that show the expression at two discontinuous developmental stages. Gene names for the ApiAp2 domain containing proteins are given by the sides, and an arrow next to the gene name indicates the presence of an ortholog in Cryposporidium. Note that there is at least one TF from each stage that has an ortholog in Cryptosporidium. The graphs on the right represent the average expression profile of non-ApiAp2 genes that show a high correlation in their expression profile with the ApiAp2 genes. The expression of such genes in a stage-specific manner suggests that these genes could be the potential targets for the predicted TFs.
Similar articles
- The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems.
Oberstaller J, Pumpalova Y, Schieler A, Llinás M, Kissinger JC. Oberstaller J, et al. Nucleic Acids Res. 2014 Jul;42(13):8271-84. doi: 10.1093/nar/gku500. Epub 2014 Jun 23. Nucleic Acids Res. 2014. PMID: 24957599 Free PMC article. - Phylogeny and domain evolution in the APETALA2-like gene family.
Kim S, Soltis PS, Wall K, Soltis DE. Kim S, et al. Mol Biol Evol. 2006 Jan;23(1):107-20. doi: 10.1093/molbev/msj014. Epub 2005 Sep 8. Mol Biol Evol. 2006. PMID: 16151182 - From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants.
Magnani E, Sjölander K, Hake S. Magnani E, et al. Plant Cell. 2004 Sep;16(9):2265-77. doi: 10.1105/tpc.104.023135. Epub 2004 Aug 19. Plant Cell. 2004. PMID: 15319480 Free PMC article. - DNA-binding domains of plant-specific transcription factors: structure, function, and evolution.
Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S. Yamasaki K, et al. Trends Plant Sci. 2013 May;18(5):267-76. doi: 10.1016/j.tplants.2012.09.001. Epub 2012 Oct 3. Trends Plant Sci. 2013. PMID: 23040085 Review. - Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes.
Iyer LM, Anantharaman V, Wolf MY, Aravind L. Iyer LM, et al. Int J Parasitol. 2008 Jan;38(1):1-31. doi: 10.1016/j.ijpara.2007.07.018. Epub 2007 Sep 15. Int J Parasitol. 2008. PMID: 17949725 Review.
Cited by
- Optogenetic modulation of an adenylate cyclase in Toxoplasma gondii demonstrates a requirement of the parasite cAMP for host-cell invasion and stage differentiation.
Hartmann A, Arroyo-Olarte RD, Imkeller K, Hegemann P, Lucius R, Gupta N. Hartmann A, et al. J Biol Chem. 2013 May 10;288(19):13705-17. doi: 10.1074/jbc.M113.465583. Epub 2013 Mar 22. J Biol Chem. 2013. PMID: 23525100 Free PMC article. - The unfolded protein response in the protozoan parasite Toxoplasma gondii features translational and transcriptional control.
Joyce BR, Tampaki Z, Kim K, Wek RC, Sullivan WJ Jr. Joyce BR, et al. Eukaryot Cell. 2013 Jul;12(7):979-89. doi: 10.1128/EC.00021-13. Epub 2013 May 10. Eukaryot Cell. 2013. PMID: 23666622 Free PMC article. - Localization in vivo and in vitro confirms EnApiAP2 protein encoded by ENH_00027130 as a nuclear protein in Eimeria necatrix.
Cai W, Feng Q, Wang L, Su S, Hou Z, Liu D, Kang X, Xu J, Pan Z, Tao J. Cai W, et al. Front Cell Infect Microbiol. 2023 Dec 5;13:1305727. doi: 10.3389/fcimb.2023.1305727. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38116134 Free PMC article. - Comparison of high throughput RNA sequences between Babesia bigemina and Babesia bovis revealed consistent differential gene expression that is required for the Babesia life cycle in the vertebrate and invertebrate hosts.
Capelli-Peixoto J, Saelao P, Johnson WC, Kappmeyer L, Reif KE, Masterson HE, Taus NS, Suarez CE, Brayton KA, Ueti MW. Capelli-Peixoto J, et al. Front Cell Infect Microbiol. 2022 Dec 19;12:1093338. doi: 10.3389/fcimb.2022.1093338. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36601308 Free PMC article. - Transcriptomic analysis reveals reduced transcriptional activity in the malaria parasite Plasmodium cynomolgi during progression into dormancy.
Bertschi NL, Voorberg-van der Wel A, Zeeman AM, Schuierer S, Nigsch F, Carbone W, Knehr J, Gupta DK, Hofman SO, van der Werff N, Nieuwenhuis I, Klooster E, Faber BW, Flannery EL, Mikolajczak SA, Chuenchob V, Shrestha B, Beibel M, Bouwmeester T, Kangwanrangsan N, Sattabongkot J, Diagana TT, Kocken CH, Roma G. Bertschi NL, et al. Elife. 2018 Dec 27;7:e41081. doi: 10.7554/eLife.41081. Elife. 2018. PMID: 30589413 Free PMC article.
References
- Lodish H., Berk A., Zipursky S.L., Matsudaira P., Baltimore D., Darnell J.E. Molecular Cell Biology. NY: W.H. Freeman & Co.; 1999.
- Cramer P. Common structural features of nucleic acid polymerases. Bioessays. 2002;24:724–729. - PubMed
- Borukhov S., Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr. Opin. Microbiol. 2003;6:93–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous