Sensitive sequencing method for KRAS mutation detection by Pyrosequencing - PubMed (original) (raw)
Comparative Study
Sensitive sequencing method for KRAS mutation detection by Pyrosequencing
Shuji Ogino et al. J Mol Diagn. 2005 Aug.
Abstract
Both benign and malignant tumors represent heterogenous tissue containing tumor cells and non-neoplastic mesenchymal and inflammatory cells. To detect a minority of mutant KRAS alleles among abundant wild-type alleles, we developed a sensitive DNA sequencing assay using Pyrosequencing, ie, nucleotide extension sequencing with an allele quantification capability. We designed our Pyrosequencing assay for use with whole-genome-amplified DNA from paraffin-embedded tissue. Assessing various mixtures of DNA from mutant KRAS cell lines and DNA from a wild-type KRAS cell line, we found that mutation detection rates for Pyrosequencing were superior to dideoxy sequencing. In addition, Pyrosequencing proved superior to dideoxy sequencing in the detection of KRAS mutations from DNA mixtures of paraffin-embedded colon cancer and normal tissue as well as from paraffin-embedded pancreatic cancers. Quantification of mutant alleles by Pyrosequencing was precise and useful for assay validation, monitoring, and quality assurance. Our Pyrosequencing method is simple, robust, and sensitive, with a detection limit of approximately 5% mutant alleles. It is particularly useful for tumors containing abundant non-neoplastic cells. In addition, the applicability of this assay for DNA amplified by whole-genome amplification technique provides an expanded source of DNA for large-scale studies.
Figures
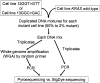
Figure 1
KRAS Pyrosequencing assay design. A: PCR amplifies a segment of KRAS exon 1 containing codons 12 and 13. The reverse PCR primer is biotinylated. B: KRAS-PF1 Pyrosequencing primer can detect common codon 12 mutations. C: KRAS-PF2 Pyrosequencing primer can detect the rest of the codon 12 mutations. D: KRAS-PF3 Pyrosequencing primer can detect codon 13 mutations. WT, wild type; Mut, mutant.
Figure 2
Pyrograms of wild-type and mutant KRAS. A: Wild-type codon 12 by the KRAS-PF1 primer. B: c.35G>T (codon 12 GTT) mutation by the KRAS-PF1 primer. C: c.35G>A (codon 12 GAT) mutation by the KRAS-PF1 primer. D: Wild-type codon 12 by the KRAS-PF2 primer. E: c.34G>T (codon 12 TGT) mutation by the KRAS-PF2 primer. F: c.34G>A (codon 12 AGT) mutation by the KRAS-PF2 primer. G: Wild-type codon 13 by the KRAS-PF3 primer. H: c.38G>A (codon 13 GAC) mutation by the KRAS-PF3 primer. Arrows indicate the presence of mutant alleles. Note that B, C, E, F, and H show the presence of wild-type sequence, which is derived from non-neoplastic cells in tumor (and a normal allele in tumor cells).
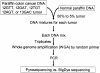
Figure 3
Cell line DNA mixing study.
Figure 4
Paraffin colon cancer DNA mixing study.
Figure 5
Allele quantifications by KRAS Pyrosequencing. The horizontal axis shows expected value of the fraction of mutant KRAS based on various cell line DNA mixtures. The vertical axis shows observed quantification results by Pyrosequencing. “Regular DNA” indicates results without WGA. The vertical bar from each dot indicates ±1 SD.
Figure 6
Comparison of Pyrosequencing and BigDye Terminator sequencing. A: Cell line DNA mixing study. A1: The c.35G>T (codon 12 GTT) mutation can be observed by both methods. A2: The c.35G>T (codon 12 GTT) mutation in the 2% mix can easily be observed by Pyrosequencing (left) but is subtle in BigDye sequencing (right). A3 and A4: The c.38G>A (codon 13 GAC) mutation can barely be observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right). B: Paraffin colon tumor DNA mixing study. B1 and B2: The c.35G>T (codon 12 GTT) mutation can be barely observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right). B3 and B4: The c.35G>A (codon 12 GAT) mutation can barely be observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right). B5 and B6: The c.34G>A (codon 12 AGT) mutation can readily be observed by Pyrosequencing (left) but cannot be detected by BigDye sequencing (right).
Similar articles
- Pyrosequencing method to detect KRAS mutation in formalin-fixed and paraffin-embedded tumor tissues.
Dufort S, Richard MJ, de Fraipont F. Dufort S, et al. Anal Biochem. 2009 Aug 15;391(2):166-8. doi: 10.1016/j.ab.2009.05.027. Epub 2009 May 21. Anal Biochem. 2009. PMID: 19464247 - KRAS detection in colonic tumors by DNA extraction from FTA paper: the molecular touch-prep.
Petras ML, Lefferts JA, Ward BP, Suriawinata AA, Tsongalis GJ. Petras ML, et al. Diagn Mol Pathol. 2011 Dec;20(4):189-93. doi: 10.1097/PDM.0b013e318211d554. Diagn Mol Pathol. 2011. PMID: 22089345 - Detection of K-ras mutations in pancreatic and hepatic neoplasms by non-isotopic mismatched polymerase chain reaction.
Stork P, Loda M, Bosari S, Wiley B, Poppenhusen K, Wolfe H. Stork P, et al. Oncogene. 1991 May;6(5):857-62. Oncogene. 1991. PMID: 1646989 - Development of pyrosequencing methods for the rapid detection of RAS mutations in clinical samples.
Cortes U, Guilloteau K, Rouvreau M, Archaimbault C, Villalva C, Karayan-Tapon L. Cortes U, et al. Exp Mol Pathol. 2015 Oct;99(2):207-11. doi: 10.1016/j.yexmp.2015.07.003. Epub 2015 Jul 8. Exp Mol Pathol. 2015. PMID: 26163758
Cited by
- Frequency of TP53 Mutations and its Impact on Drug Sensitivity in Acute Myeloid Leukemia?
Shah A, Seedhouse C. Shah A, et al. Indian J Clin Biochem. 2012 Apr;27(2):121-6. doi: 10.1007/s12291-012-0203-1. Epub 2012 Mar 24. Indian J Clin Biochem. 2012. PMID: 23543587 Free PMC article. Retracted. - Up-regulation of c-MYC and SIRT1 expression correlates with malignant transformation in the serrated route to colorectal cancer.
Kriegl L, Vieth M, Kirchner T, Menssen A. Kriegl L, et al. Oncotarget. 2012 Oct;3(10):1182-93. doi: 10.18632/oncotarget.628. Oncotarget. 2012. PMID: 23045412 Free PMC article. - Associations of artificially sweetened beverage intake with disease recurrence and mortality in stage III colon cancer: Results from CALGB 89803 (Alliance).
Guercio BJ, Zhang S, Niedzwiecki D, Li Y, Babic A, Morales-Oyarvide V, Saltz LB, Mayer RJ, Mowat RB, Whittom R, Hantel A, Benson A, Atienza D, Messino M, Kindler H, Venook A, Ogino S, Zoltick ES, Stampfer M, Ng K, Wu K, Willett WC, Giovannucci EL, Meyerhardt JA, Fuchs CS. Guercio BJ, et al. PLoS One. 2018 Jul 19;13(7):e0199244. doi: 10.1371/journal.pone.0199244. eCollection 2018. PLoS One. 2018. PMID: 30024889 Free PMC article. - IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53.
Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS, Ogino S. Kawasaki T, et al. Neoplasia. 2007 Dec;9(12):1091-8. doi: 10.1593/neo.07760. Neoplasia. 2007. PMID: 18084616 Free PMC article. - Predictive and prognostic analysis of PIK3CA mutation in stage III colon cancer intergroup trial.
Ogino S, Liao X, Imamura Y, Yamauchi M, McCleary NJ, Ng K, Niedzwiecki D, Saltz LB, Mayer RJ, Whittom R, Hantel A, Benson AB 3rd, Mowat RB, Spiegelman D, Goldberg RM, Bertagnolli MM, Meyerhardt JA, Fuchs CS; Alliance for Clinical Trials in Oncology. Ogino S, et al. J Natl Cancer Inst. 2013 Dec 4;105(23):1789-98. doi: 10.1093/jnci/djt298. Epub 2013 Nov 14. J Natl Cancer Inst. 2013. PMID: 24231454 Free PMC article. Clinical Trial.
References
- Fakhrai-Rad H, Pourmand N, Ronaghi M. Pyrosequencing: an accurate detection platform for single nucleotide polymorphisms. Hum Mutat. 2002;19:479–485. - PubMed
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. - PubMed
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous