Generating chromosome instability through the simultaneous deletion of Mad2 and p53 - PubMed (original) (raw)
Comparative Study
. 2005 Aug 9;102(32):11296-301.
doi: 10.1073/pnas.0505053102. Epub 2005 Jul 29.
Affiliations
- PMID: 16055552
- PMCID: PMC1182134
- DOI: 10.1073/pnas.0505053102
Comparative Study
Generating chromosome instability through the simultaneous deletion of Mad2 and p53
Aurora A Burds et al. Proc Natl Acad Sci U S A. 2005.
Abstract
Cancer cells exhibit high levels of chromosome instability (CIN), and considerable interest surrounds the possibility that inactivation of the spindle checkpoint is involved. However, homozygous disruption of Mad and Bub checkpoint genes in metazoans causes cell death rather than CIN. We now report the isolation and characterization of blastocysts and two independent mouse embryonic fibroblast lines carrying deletions in Mad2 and p53. These cells lack a functional spindle checkpoint, undergo anaphase prematurely, and exhibit an extraordinarily high level of CIN. We conclude that the mitotic checkpoint is not essential for viability per se and that a CIN phenotype can be established in culture through the inactivation of both the Mad2- and p53-dependent checkpoint pathways.
Figures
Fig. 1.
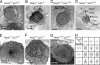
Characterization of Mad2_–/– blastocysts. Blastocysts of Mad2+/– p53+/_+ intercrosses or Mad2+/– _p53_–/– × Mad2+/– p53+/– matings were isolated at E3.5. The growth of blastocysts was recorded on a daily basis, and blastocysts were harvested for PCR genotyping when death appeared imminent. Cultured blastocysts were imaged before harvest by using phase-contrast microscopy 4–5 days (A_–_D) or 28 days (E_–_G) after isolation. Arrows indicate inner cell mass and trophoblast giant cells. (H) The genotypes of 102 recovered blastocysts are indicated. Ratios indicate how many of the genotyped blastocysts appeared healthy when harvested for PCR.
Fig. 2.
Generation and characterization of Mad2_–/– p53_–/– MEF cell lines. (A) Numbers of E10.5 embryos with the indicated genotypes. Embryos from natural matings were recovered at E10.5. (B) Genotypes of MEF cell lines. (C) MEF DNA was subjected to PCR at the Mad2 (lanes 1–4) and p53 loci (lanes 5 and 6). Mad2+/+ _p53_–/– (AB103), Mad2+/– _p53_–/– (AB100) and _Mad2_–/– _p53_–/– (AB98) cell lines were derived from embryos of the same litter. The bands for mutant and wild-type alleles are indicated by “m” and “w,” respectively. Y-sacAB152, genotyping of the yolk sac of AB152. (D) MEFs were grown in culture and imaged to compare cell morphology and growth. Shown are phase-contrast images (Upper) and, at higher magnification, phase-contrast/fluorescence images (Lower) taken of cells expressing H2B–GFP to visualize chromatin. Images were taken with a ×10, 0.4 numerical aperture objective. (Scale bars: Upper, 100 μm; Lower, 10 μm.) (E) Mad2 mRNA levels as assayed by RT-PCR. Total RNA from AB103, AB100, and AB98 MEF lines was isolated, transcribed into cDNA, and subjected to PCR by using _Mad2_-specific (lanes 1–3) and _GAPDH_-specific (lanes 3–5) oligonucleotides. (F) Mad2 and γ-tubulin levels in lysates of 3T3 cells, AB103, AB100, and AB98 MEF lines were determined by using Western blotting; 2.5 ng of recombinant mouse Mad2 protein (lane 1) was loaded as a positive control. The Mad2 polyclonal antibody recognizes an uncharacterized band (*) in 3T3 cells but not in other MEFs that electrophoreses more slowly than the Mad2 band. (G) Checkpoint status as monitored by live cell imaging. AB103 and AB98 expressing H2B–GFP were treated with 330 nM nocodazole and imaged by time-lapse microscopy. The graph shows the percentage of cells that have undergone anaphase B and exited mitosis 60 min after NBD.
Fig. 3.
Live cell imaging and accelerated mitotic progression in cells lacking Mad2. (A_–_C) Images of Mad2+/+ _p53_–/– (AB103) and _Mad2_–/– _p53_–/– (AB98) during mitosis by using time-lapse microscopy. Chromatin was visualized with H2B–GFP. The T = 0 min time point shows cells shortly before NBD, as judged by loss of nuclear integrity and compact organization of the chromosomes. Cells were imaged every 2 min from this point until chromosome decondensation after anaphase. Pictures were taken every 2 min using a ×63 objective on a DeltaVision microscope (Zeiss). (Scale bars: 10 μm.) M indicates a lagging chromosome that seems to form a micronucleus in anaphase. (D) Quantitation of data from live cell imaging. Images were taken every 4 min with a ×20 objective. The time between NBD and anaphase onset was determined for at least 30 cells and is shown as accumulative graph. Mad2_–/– p53_–/– cell lines AB98 and AB152 (shown as the average of both cell lines) show accelerated progression although mitosis, compared with Mad2+/+ _p53_–/– (AB103) and NIH 3T3 cells. (E) Summary of mitotic timing data presented in D. In each case, the mode of the time of anaphase onset is shown. The error in mitotic times was estimated by subsampling the data and determining the standard deviation in the mode; n shows the total number of cells that were analyzed.
Fig. 4.
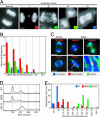
Chromosome missegregation in Mad2_–/– MEFs. (A and B) Images of DAPI-stained cells sorted into categories of increasingly severe chromosome mis-segregation, as judged visually. (Scale bars: 5 μm.) The extent of missegregation was then quantified in each cell line (B) by using the classification shown in A. (C) Immunostaining (chromatin in blue, tubulin in green, and BubR1/Bub1 in red) of Mad2+/_+ _p53_–/– (AB103) and _Mad2_–/– _p53_–/– (AB98) cells. Arrows point to lagging chromosomes that stain positive for BubR1 or Bub1. A white box marks a chromosome that appears to have merotelic attachment to microtubules emanating from both poles. The boxed region was enhanced to present 3D information more clearly by using
volocity
software (Improvision, Lexington, MA) and is shown magnified in the Lower Right. (Scale bars:10 μm, except for 1 μminthe Lower Right.) (D) Diploid and tetraploid states of wild-type MEFs and _Mad2_–/– _p53_–/– (AB98) and AB152 MEF cell lines as determined by FACS analysis. (E) Diploid and tetraploid states of AB98 and AB152, respectively, as determined by karyotyping. The graph shows the distribution of chromosome numbers in metaphase spreads.
Fig. 5.
Cycling Mad2_–/– p53_–/– MEFs do not accumulate high levels of double-strand breaks. (A) Indirect immunofluorescence of phospho-H2AX. Mad2+/+ _p53_–/– (AB103) and _Mad2_–/– _p53_–/– (AB98) cells were irradiated with 1,700 rad in a γ cell irradiator. Cells were fixed and stained 30 min after irradiation. –γ, untreated cells; +γ, irradiated cells; DAPI, blue; phospho-H2AX, green. (Scale bar: 10 μm.) Percentage of phospho-H2AX-positive cells is indicated in the images. (B) Quantitation of phospho-H2AX levels in lysates from NIH 3T3 cells, and AB103, AB100, and AB98 MEF lines before and after irradiation. Lysates were prepared from untreated cells and from cells 1 h after irradiation. Immunoblots were probed by using phosphospecific H2AX antibody and monoclonal γ-tubulin antibody as loading control. Phospho-H2AX protein levels were quantitated and normalized to γ-tubulin protein levels by using a FluoroImager459 (Molecular Dynamics) and
imagequant
software (Molecular Dynamics).
Similar articles
- Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition.
Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Schvartzman JM, et al. Cancer Cell. 2011 Jun 14;19(6):701-14. doi: 10.1016/j.ccr.2011.04.017. Cancer Cell. 2011. PMID: 21665145 Free PMC article. - MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells.
Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. Michel LS, et al. Nature. 2001 Jan 18;409(6818):355-9. doi: 10.1038/35053094. Nature. 2001. PMID: 11201745 - Whole chromosome instability resulting from the synergistic effects of pRB and p53 inactivation.
Manning AL, Benes C, Dyson NJ. Manning AL, et al. Oncogene. 2014 May 8;33(19):2487-94. doi: 10.1038/onc.2013.201. Epub 2013 Jun 24. Oncogene. 2014. PMID: 23792446 Free PMC article. - The Mad1-Mad2 balancing act--a damaged spindle checkpoint in chromosome instability and cancer.
Schuyler SC, Wu YF, Kuan VJ. Schuyler SC, et al. J Cell Sci. 2012 Sep 15;125(Pt 18):4197-206. doi: 10.1242/jcs.107037. Epub 2012 Oct 23. J Cell Sci. 2012. PMID: 23093575 Review. - The mitotic checkpoint: a signaling pathway that allows a single unattached kinetochore to inhibit mitotic exit.
Chan GK, Yen TJ. Chan GK, et al. Prog Cell Cycle Res. 2003;5:431-9. Prog Cell Cycle Res. 2003. PMID: 14593737 Review.
Cited by
- Linking stem cells to chromosomal instability.
van Wely KH, Martínez-A C. van Wely KH, et al. Oncoimmunology. 2012 Mar 1;1(2):195-200. doi: 10.4161/onci.1.2.18613. Oncoimmunology. 2012. PMID: 22720241 Free PMC article. - Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice.
Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Baker DJ, et al. J Cell Biol. 2006 Feb 13;172(4):529-40. doi: 10.1083/jcb.200507081. J Cell Biol. 2006. PMID: 16476774 Free PMC article. - Inactivating the spindle checkpoint kinase Bub1 during embryonic development results in a global shutdown of proliferation.
Tilston V, Taylor SS, Perera D. Tilston V, et al. BMC Res Notes. 2009 Sep 23;2:190. doi: 10.1186/1756-0500-2-190. BMC Res Notes. 2009. PMID: 19772675 Free PMC article. - Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Kwiatkowski N, Jelluma N, Filippakopoulos P, Soundararajan M, Manak MS, Kwon M, Choi HG, Sim T, Deveraux QL, Rottmann S, Pellman D, Shah JV, Kops GJ, Knapp S, Gray NS. Kwiatkowski N, et al. Nat Chem Biol. 2010 May;6(5):359-68. doi: 10.1038/nchembio.345. Epub 2010 Apr 11. Nat Chem Biol. 2010. PMID: 20383151 Free PMC article. - Studying chromosome instability in the mouse.
Foijer F, Draviam VM, Sorger PK. Foijer F, et al. Biochim Biophys Acta. 2008 Sep;1786(1):73-82. doi: 10.1016/j.bbcan.2008.07.004. Epub 2008 Jul 26. Biochim Biophys Acta. 2008. PMID: 18706976 Free PMC article. Review.
References
- Boveri, T. (1914) Zur frage der entstehung maligner tumoren (Fischer, Jena, Germany); trans. Boveri, M. (1929) The Origin of Malignant Tumours (Williams & Wilkins, Baltimore).
- Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1997) Nature 386, 623–627. - PubMed
- Draviam, V. M., Xie, S. & Sorger, P. K. (2004) Curr. Opin. Genet. Dev. 14, 120–125. - PubMed
- Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112, 407–421. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous