miBLAST: scalable evaluation of a batch of nucleotide sequence queries with BLAST - PubMed (original) (raw)
miBLAST: scalable evaluation of a batch of nucleotide sequence queries with BLAST
You Jung Kim et al. Nucleic Acids Res. 2005.
Abstract
A common task in many modern bioinformatics applications is to match a set of nucleotide query sequences against a large sequence dataset. Existing tools, such as BLAST, are designed to evaluate a single query at a time and can be unacceptably slow when the number of sequences in the query set is large. In this paper, we present a new algorithm, called miBLAST, that evaluates such batch workloads efficiently. At the core, miBLAST employs a q-gram filtering and an index join for efficiently detecting similarity between the query sequences and database sequences. This set-oriented technique, which indexes both the query and the database sets, results in substantial performance improvements over existing methods. Our results show that miBLAST is significantly faster than BLAST in many cases. For example, miBLAST aligned 247 965 oligonucleotide sequences in the Affymetrix probe set against the Human UniGene in 1.26 days, compared with 27.27 days with BLAST (an improvement by a factor of 22). The relative performance of miBLAST increases for larger word sizes; however, it decreases for longer queries. miBLAST employs the familiar BLAST statistical model and output format, guaranteeing the same accuracy as BLAST and facilitating a seamless transition for existing BLAST users.
Figures
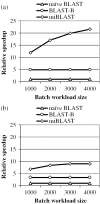
Figure 1
This graph shows the relative speedup of each method compared with naive BLAST, for various workload sizes using a word size of 11. (a) Affymetrix (25 bp). (b) Illumina (70 bp).
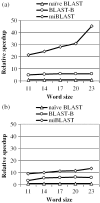
Figure 2
This graph shows the effect of the BLAST word size parameter on the query performance for each method, plotted as relative speedup over naive BLAST. The batch size used in this experiment is 4000 queries. miBLAST uses an index word size of 11 and uses a sliding-window filtering method for query word sizes between 14 and 23. (a) Affymetrix (25 bp). (b) Illumina (70 bp).
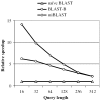
Figure 3
This graph shows the relative speedup to naive BLAST for various query lengths, using a word size of 11. Queries are drawn from the EST human dataset, and each batch has 1000 queries.
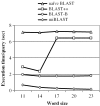
Figure 4
This graph shows the execution time of each method for various word sizes using a batch of 1000 queries from the Affymetrix probe set (25 bp).
Similar articles
- Parallelisation of the blast algorithm.
Qi Y, Lin F. Qi Y, et al. Cell Mol Biol Lett. 2005;10(2):281-5. Cell Mol Biol Lett. 2005. PMID: 16010293 - BOV--a web-based BLAST output visualization tool.
Gollapudi R, Revanna KV, Hemmerich C, Schaack S, Dong Q. Gollapudi R, et al. BMC Genomics. 2008 Sep 15;9:414. doi: 10.1186/1471-2164-9-414. BMC Genomics. 2008. PMID: 18793422 Free PMC article. - NdPASA: a novel pairwise protein sequence alignment algorithm that incorporates neighbor-dependent amino acid propensities.
Wang J, Feng JA. Wang J, et al. Proteins. 2005 Feb 15;58(3):628-37. doi: 10.1002/prot.20359. Proteins. 2005. PMID: 15616964 - Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre.
Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Bennett-Lovsey RM, et al. Proteins. 2008 Feb 15;70(3):611-25. doi: 10.1002/prot.21688. Proteins. 2008. PMID: 17876813 - High speed BLASTN: an accelerated MegaBLAST search tool.
Chen Y, Ye W, Zhang Y, Xu Y. Chen Y, et al. Nucleic Acids Res. 2015 Sep 18;43(16):7762-8. doi: 10.1093/nar/gkv784. Epub 2015 Aug 6. Nucleic Acids Res. 2015. PMID: 26250111 Free PMC article.
Cited by
- BatchGenAna: a batch platform for large-scale genomic analysis of mammalian small RNAs.
Ying X, Kim YJ, Mao Y, Liu M, Hou Y, Li H, Wang X, Zhao Y, Zhao D, Patel JM, Li W. Ying X, et al. Bioinformation. 2009 Apr 21;3(8):346-8. doi: 10.6026/97320630003346. Bioinformation. 2009. PMID: 19707298 Free PMC article. - Integrating in silico resources to map a signaling network.
Liu H, Beck TN, Golemis EA, Serebriiskii IG. Liu H, et al. Methods Mol Biol. 2014;1101:197-245. doi: 10.1007/978-1-62703-721-1_11. Methods Mol Biol. 2014. PMID: 24233784 Free PMC article. - Applications of the pipeline environment for visual informatics and genomics computations.
Dinov ID, Torri F, Macciardi F, Petrosyan P, Liu Z, Zamanyan A, Eggert P, Pierce J, Genco A, Knowles JA, Clark AP, Van Horn JD, Ames J, Kesselman C, Toga AW. Dinov ID, et al. BMC Bioinformatics. 2011 Jul 26;12:304. doi: 10.1186/1471-2105-12-304. BMC Bioinformatics. 2011. PMID: 21791102 Free PMC article. - Simple Matching Using QIIME 2 and RDP Reveals Misidentified Sequences and an Underrepresentation of Fungi in Reference Datasets.
Eldred LE, Thorn RG, Smith DR. Eldred LE, et al. Front Genet. 2021 Nov 26;12:768473. doi: 10.3389/fgene.2021.768473. eCollection 2021. Front Genet. 2021. PMID: 34899856 Free PMC article. - Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together.
Jayapandian M, Chapman A, Tarcea VG, Yu C, Elkiss A, Ianni A, Liu B, Nandi A, Santos C, Andrews P, Athey B, States D, Jagadish HV. Jayapandian M, et al. Nucleic Acids Res. 2007 Jan;35(Database issue):D566-71. doi: 10.1093/nar/gkl859. Epub 2006 Nov 27. Nucleic Acids Res. 2007. PMID: 17130145 Free PMC article.
References
- Lachance P.E., Chaudhuri A. Microaarry analysis of developmental plasticity in monkey primary visual cortex. J. Neurochem. 2004;88:1455–1469. - PubMed
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
- Altschul S., Gish W. Local alignment statistics. Meth. Enzymol. 1996;266:460–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials