The origin of the naked grains of maize - PubMed (original) (raw)
The origin of the naked grains of maize
Huai Wang et al. Nature. 2005.
Abstract
The most critical step in maize (Zea mays ssp. mays) domestication was the liberation of the kernel from the hardened, protective casing that envelops the kernel in the maize progenitor, teosinte. This evolutionary step exposed the kernel on the surface of the ear, such that it could readily be used by humans as a food source. Here we show that this key event in maize domestication is controlled by a single gene (teosinte glume architecture or tga1), belonging to the SBP-domain family of transcriptional regulators. The factor controlling the phenotypic difference between maize and teosinte maps to a 1-kilobase region, within which maize and teosinte show only seven fixed differences in their DNA sequences. One of these differences encodes a non-conservative amino acid substitution and may affect protein function, and the other six differences potentially affect gene regulation. Molecular evolution analyses show that this region was the target of selection during maize domestication. Our results demonstrate that modest genetic changes in single genes can induce dramatic changes in phenotype during domestication and evolution.
Figures
Figure 1
Phenotypes. a. Maize ear showing the cob (cb) exposed at top. b. Teosinte ear with the rachis internode (in) and glume (gl) labeled. c. Teosinte ear from a plant with a maize allele of tga1 introgressed into it. d. Close-up of a single teosinte fruitcase. e. Close-up of a fruitcase from teosinte plant with a maize allele of tga1 introgressed into it. f. Ear of maize inbred W22 (Tga1-maize allele) with the cob exposed showing the small white glumes at the base. g. Ear of maize inbred W22:tga1 which carries the teosinte allele, showing enlarged (white) glumes. h. Ear of maize inbred W22 carrying the tga1-ems1 allele, showing enlarged glumes. For higher magnification copies of f–h see Supplementary Information.
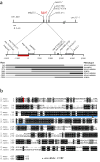
Figure 2
The tga1 locus. a. Map of the chromosomal segment at tga1 with marker loci used for positional cloning. Hatched boxes are the three exons of tga1. Red box is the 1042 bp segment to which the factor controlling the difference between maize and teosinte was mapped. Seven of the 3106 plants with cross-overs in tga1 are shown along with their phenotypes and genotypes (light gray bar: homozygous maize; medium gray bar: heterozygous; black bar: homozygous teosinte). For details on marker loci see supplemental information. b. The tga1 protein sequence aligned with its rice counterpart. Identical amino acids are in black background and similar ones in gray. Blue bar indicates the SBP domain, green triangles indicate the intron positions. The fixed amino acid difference between teosinte and maize (K→N) is highlighted in red. The position of the L→F amino acid change for the tga1-ems1 allele is marked with an asterisk.
Figure 3
Molecular analysis of tga1. a. Top panel is a Northern blot showing that tga1 is expressed in immature ears of W22 maize (M) and W22:tga1 (T) which carries the teosinte allele. Weak expression is also visible in the husks (HK). No expression of tga1 is seen in root (RT), prop root (PR), unexpanded leaf (LF) or immature tassel (TS). Lower panel is the ethidium bromide stained gel with the visible rRNAs confirming approximately equal loading of total RNA in each lane. b. Western blots showing that the protein encoded by the teosinte allele of tga1 accumulates at a higher level than the protein of the maize allele. Blots probed with either anti-TGA1 or anti-ACTIN (control) antibodies. Protein was extracted from individual ears of W22 maize (M) carrying a maize allele and W22:tga1 (T) carrying a teosinte allele. Immature ears were staged by their lengths which are indicated in mm.
Figure 4
Tissue in situ hybridizations. a–d, Immature ears. e–h, Young spikelet primordia. i–l, Older spikelet primordia with distinct floral organ primordia. tga1 antisense probe was applied to W22 maize (a, e, i), W22:tga1 (b, f, j), and teosinte (c, g, k). tga1 sense probe was applied to W22 as a control (d, h, l). Arrows show tga1 expression in the inflorescence meristem (a), spikelet-pair primordium (b), teosinte husk (c), glume (e), and cupule forming region (i).
Figure 5
Molecular evolution. a, Nucleotide diversity (π) in maize and teosinte for tga1. Tajima’s D and HKA tests for non-neutral evolution and the ratio of π in maize to π in teosinte are shown. b, Joint distribution of the posterior probabilities (scale from white to black) for the time since fixation of the maize allele in generation (T gen) and the selection coefficient (s).
Similar articles
- Evidence That the Origin of Naked Kernels During Maize Domestication Was Caused by a Single Amino Acid Substitution in tga1.
Wang H, Studer AJ, Zhao Q, Meeley R, Doebley JF. Wang H, et al. Genetics. 2015 Jul;200(3):965-74. doi: 10.1534/genetics.115.175752. Epub 2015 May 4. Genetics. 2015. PMID: 25943393 Free PMC article. - The role of teosinte glume architecture (tga1) in coordinated regulation and evolution of grass glumes and inflorescence axes.
Preston JC, Wang H, Kursel L, Doebley J, Kellogg EA. Preston JC, et al. New Phytol. 2012 Jan;193(1):204-215. doi: 10.1111/j.1469-8137.2011.03908.x. Epub 2011 Sep 29. New Phytol. 2012. PMID: 21954998 - The evolution of apical dominance in maize.
Doebley J, Stec A, Hubbard L. Doebley J, et al. Nature. 1997 Apr 3;386(6624):485-8. doi: 10.1038/386485a0. Nature. 1997. PMID: 9087405 - The genetics of maize evolution.
Doebley J. Doebley J. Annu Rev Genet. 2004;38:37-59. doi: 10.1146/annurev.genet.38.072902.092425. Annu Rev Genet. 2004. PMID: 15568971 Review. - Genetic, evolutionary and plant breeding insights from the domestication of maize.
Hake S, Ross-Ibarra J. Hake S, et al. Elife. 2015 Mar 25;4:e05861. doi: 10.7554/eLife.05861. Elife. 2015. PMID: 25807085 Free PMC article. Review.
Cited by
- Functional Evolution in the Plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family.
Preston JC, Hileman LC. Preston JC, et al. Front Plant Sci. 2013 Apr 5;4:80. doi: 10.3389/fpls.2013.00080. eCollection 2013. Front Plant Sci. 2013. PMID: 23577017 Free PMC article. - A natural mutation of the NST1 gene arrests secondary cell wall biosynthesis in the seed coat of a hull-less pumpkin accession.
Lyu X, Shi L, Zhao M, Li Z, Liao N, Meng Y, Ma Y, Zhou Y, Xue Q, Hu Z, Yang J, Zhang M. Lyu X, et al. Hortic Res. 2022 Jun 16;9:uhac136. doi: 10.1093/hr/uhac136. eCollection 2022. Hortic Res. 2022. PMID: 36072840 Free PMC article. - Expanding Gene-Editing Potential in Crop Improvement with Pangenomes.
Tay Fernandez CG, Nestor BJ, Danilevicz MF, Marsh JI, Petereit J, Bayer PE, Batley J, Edwards D. Tay Fernandez CG, et al. Int J Mol Sci. 2022 Feb 18;23(4):2276. doi: 10.3390/ijms23042276. Int J Mol Sci. 2022. PMID: 35216392 Free PMC article. Review. - SBP-domain transcription factors as possible effectors of cryptochrome-mediated blue light signalling in the moss Physcomitrella patens.
Riese M, Zobell O, Saedler H, Huijser P. Riese M, et al. Planta. 2008 Jan;227(2):505-15. doi: 10.1007/s00425-007-0661-5. Epub 2007 Nov 8. Planta. 2008. PMID: 17989994 Free PMC article. - Architectural evolution and its implications for domestication in grasses.
Doust A. Doust A. Ann Bot. 2007 Nov;100(5):941-50. doi: 10.1093/aob/mcm040. Epub 2007 May 3. Ann Bot. 2007. PMID: 17478546 Free PMC article. Review.
References
- Dorweiler J, Stec A, Kermicle J, Doebley J. Teosinte glume architecture1: A genetic locus controlling a key step in maize evolution. Science. 1993;262:233–235. - PubMed
- Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet. 1996;250:7–16. - PubMed
- Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38:37–59. - PubMed
- Maynard Smith J. Macroevolution. Nature. 1981;289:13–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources