High level of sequence diversity in the 16S rRNA genes of Haemophilus influenzae isolates is useful for molecular subtyping - PubMed (original) (raw)
High level of sequence diversity in the 16S rRNA genes of Haemophilus influenzae isolates is useful for molecular subtyping
Claudio T Sacchi et al. J Clin Microbiol. 2005 Aug.
Abstract
A molecular typing method based on the 16S rRNA sequence diversity was developed for Haemophilus influenzae isolates. A total of 330 H. influenzae isolates were analyzed, representing a diverse collection of U.S. isolates. We found a high level of 16S rRNA sequence heterogeneity (up to 2.73%) and observed an exclusive correlation between 16S types and serotypes (a to f); no 16S type was found in more than one serotype. Similarly, no multilocus sequence typing (MLST) sequence type (ST) was found in more than one serotype. Our 16S typing and MLST results are in agreement with those of previous studies showing that serotypable H. influenzae isolates behave as highly clonal populations and emphasize the lack of clonality of nontypable (NT) H. influenzae isolates. There was not a 1:1 correlation between 16S types and STs, but all H. influenzae serotypable isolates clustered similarly. This correlation was not observed for NT H. influenzae; the two methods clustered NT H. influenzae isolates differently. 16S rRNA gene sequencing alone provides a level of discrimination similar to that obtained with the analysis of seven genes for MLST. We demonstrated that 16S typing is an additional and complementary approach to MLST, particularly for NT H. influenzae isolates, and is potentially useful for outbreak investigation.
Figures
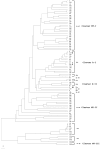
FIG.1.
Consensus UPGMA phylogenetic tree constructed with 65 identified H. influenzae 16S types. The numbers on the branches indicate the 16S type, and the respective serotypes are represented. The symbols denoting 16S types 45 (circle), 27 (open square), and 47 (filled square) indicate H. influenzae isolates with 16S types and serotypes that do not cluster with other 16S types in each respective serotype. These 16S types correlate with STs in Fig. 2, as indicated by the same symbols. The scale bar represents an expected substitution rate of 0.1 nucleotide substitution per base position. Hi-NT, nontypeable H. influenzae; Hib, H. influenzae serotype b; Hia, H. influenzae serotype a; Hic, H. influenzae serotype c; Hid, H. influenzae serotype d; Hie, H. influenzae serotype e; Hif, H. influenzae serotype f.
FIG. 2.
Consensus UPGMA phylogenetic tree constructed with 89 concatenated DNA sequences obtained from seven housekeeping genes of H. influenzae. The number in the branches indicates the ST, and the respective serotypes are represented. The symbols by STs 44 and 157 (circle), 117 (open square), and 4 (filled square) indicate STs of H. influenzae serotypes that correlate with 16S types in Fig. 1; they are indicated by the same symbols. The scale bar represents an expected substitution rate of 0.1 nucleotide substitution per base position. See the legend to Fig. 1 for abbreviations.
References
- Anonymous. 2002. Progress toward elimination of Haemophilus influenzae type b invasive disease among infants and children—United States, 1998-2000. Morb. Mortal. Wkly. Rep. 51:234-237. - PubMed
- Bujnicki, J. M. 2004. Molecular phylogenetics of restriction endonucleases, p. 63-93. In A. Pingoud (ed.), Restriction endonucleases, vol. 14. Springer-Verlag, Berlin, Germany.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials