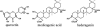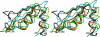Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula - PubMed (original) (raw)
Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula
Hui Shao et al. Plant Cell. 2005 Nov.
Abstract
Glycosylation is a ubiquitous reaction controlling the bioactivity and storage of plant natural products. Glycosylation of small molecules is catalyzed by a superfamily of glycosyltransferases (GTs) in most plant species studied to date. We present crystal structures of the UDP flavonoid/triterpene GT UGT71G1 from Medicago truncatula bound to UDP or UDP-glucose. The structures reveal the key residues involved in the recognition of donor substrate and, by comparison with other GT structures, suggest His-22 as the catalytic base and Asp-121 as a key residue that may assist deprotonation of the acceptor by forming an electron transfer chain with the catalytic base. Mutagenesis confirmed the roles of these key residues in donor substrate binding and enzyme activity. Our results provide an initial structural basis for understanding the complex substrate specificity and regiospecificity underlying the glycosylation of plant natural products and other small molecules. This information will direct future attempts to engineer bioactive compounds in crop plants to improve plant, animal, and human health and to facilitate the rational design of GTs to improve the storage and stability of novel engineered bioactive compounds.
Figures
Figure 1.
Structures of Three Substrates of M. truncatula UGT71G1. The known or predicted glycosylation sites are labeled with numbers: quercetin (3, 5, 7, 3′, and 4′), medicagenic acid (3 or 28), and hederagenin (3 or 28).
Figure 2.
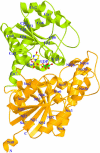
Ribbon Diagram of the Structure of UGT71G1 with Bound UDP. The N- and C-terminal domains are shown in orange and green, with the secondary structures and the N and C termini labeled. The α helices and β strands in the N- and C-terminal domains are numbered separately. The UDP molecule is shown as a ball-and-stick model colored by atom type (nitrogen, blue; carbon, yellow; oxygen, red; phosphorus, green). Figures 2, 3B, 4, 5, and 6 were prepared with MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997).
Figure 3.
Comparison of M. truncatula UGT71G1 and A. orientalis GtfD. (A) Structure-based sequence alignment of M. truncatula UGT71G1 and A. orientalis GtfD. The secondary structure elements observed in the UGT71G1 structure are shown above the alignment. The UGT signature motifs are enclosed in a green box. Conserved residues are highlighted. This figure was produced with ENDscript (Gouet and Courcelle, 2002). (B) Stereo diagram showing the superimposition of the structures of UGT71G1 (molecule A; brown) and GtfD (blue; Protein Data Bank [PDB] code 1RRV).
Figure 4.
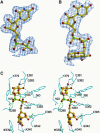
Donor Molecules and Their Interactions with UGT71G1. (A) A |F obs| − |F calc| electron density omit map of bound UDP contoured at 1.5 σ is superimposed on a ball-and-stick model of the UDP molecule. (B) A |F obs| − |F calc| electron density omit map of bound UDP-glucose contoured at 1.5 σ is superimposed on a ball-and-stick model of the UDP-glucose molecule. (C) Stereo diagram showing interactions between bound UDP-glucose and UGT71G1 side chains. The structure of UDP-glucose is shown as a ball-and-stick model. Hydrogen bonding interactions are indicated by dashed lines. Interactions observed only between UDP(-galactose) and UGT71G1 in the 2.0-Å structure are indicated by green dashed lines.
Figure 5.
Comparison of Donor Binding Regions of UGT71G1 and Other GT-B Fold Enzymes. Stereo diagram showing superimposition of the UGT signature motif regions of UGT71G1 (yellow), GtfD (red), MurG (green; PDB code 1F0K), OtsA (cyan; PDB code 1GZ5), and BGT (black; PDB code 1J39).
Figure 6.
The Putative Acceptor Binding Pocket. Stereo diagram showing quercetin (A) and hederagenin (B) docked into the proposed binding pockets. Quercetin, hederagenin, and UDP-glucose are shown as ball-and-stick models. Some protein residues in the acceptor binding pocket are labeled and shown in cyan as bond models. Distances (Å) between the OH group of acceptors and the atom NE2 of His-22 or the atom C1′ of UDP-glucose are labeled and indicated with dashed lines.
Figure 7.
Comparison of the Donor and Acceptor Regions of Functionally Characterized Plant UGTs. Sequence alignment of 39 plant UGTs, showing the acceptor binding region (first three fragments: 1 to 55, 81 to 95, and 117 to 151) and the donor binding region (last two fragments: 282 to 286 and 339 to 382). Identical residues are highlighted, and similar residues are enclosed in boxes. Residues His-22 and Asp-121 are marked with asterisks. The alignment was performed using ClustalX (Thompson et al., 1997). The plant UGTs include Medicago truncatula UGT71G1 and UGT73K1; Allium cepa UGT73G1 and UGT73J1; Aralia cordata GaT; Bellis perennis UGAT; Brassica napus SGT1; Catharanthus roseus UGT2; Citrus maxima 1,2RhaT; Crocus sativus UGTCs2; Dorotheanthus bellidiformis UGT73A5; Gentiana triflora 3′GT; Glycyrrhiza echinata UGT73F1; Nicotiana tabacum GT1a, GT2, and GT3; Phaseolus lunatus ZOG1; Phaseolus vulgaris ZOX1; Sorghum bicolor UGT85B1; Stevia rebaudiana UGT74G1, UGT76G1, and UGT85C2; and Zea mays cisZOC1 and cisZOG2. All others are from Arabidopsis thaliana.
Figure 8.
Glucosylation of Quercetin by Recombinant UGT71G1. (A) HPLC trace showing the substrate quercetin (Q) and the five products (numbered 1 to 5). Solid line, complete reaction mixture; dashed line, standards for quercetin (Q), quercetin-3-_O_-glucoside (peak 3), and quercetin-4′-_O_-glucoside (peak 4). mAU, milli-absorption unit. (B) UV absorption spectra of quercetin (Q) and products 1 to 5.
Similar articles
- Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
Modolo LV, Li L, Pan H, Blount JW, Dixon RA, Wang X. Modolo LV, et al. J Mol Biol. 2009 Oct 9;392(5):1292-302. doi: 10.1016/j.jmb.2009.08.017. Epub 2009 Aug 13. J Mol Biol. 2009. PMID: 19683002 - Mutational analysis of the Medicago glycosyltransferase UGT71G1 reveals residues that control regioselectivity for (iso)flavonoid glycosylation.
He XZ, Wang X, Dixon RA. He XZ, et al. J Biol Chem. 2006 Nov 10;281(45):34441-7. doi: 10.1074/jbc.M605767200. Epub 2006 Sep 18. J Biol Chem. 2006. PMID: 16982612 - Crystal structure of Medicago truncatula UGT85H2--insights into the structural basis of a multifunctional (iso)flavonoid glycosyltransferase.
Li L, Modolo LV, Escamilla-Trevino LL, Achnine L, Dixon RA, Wang X. Li L, et al. J Mol Biol. 2007 Jul 27;370(5):951-63. doi: 10.1016/j.jmb.2007.05.036. Epub 2007 May 18. J Mol Biol. 2007. PMID: 17553523 - Triterpenoid-biosynthetic UDP-glycosyltransferases from plants.
Rahimi S, Kim J, Mijakovic I, Jung KH, Choi G, Kim SC, Kim YJ. Rahimi S, et al. Biotechnol Adv. 2019 Nov 15;37(7):107394. doi: 10.1016/j.biotechadv.2019.04.016. Epub 2019 May 9. Biotechnol Adv. 2019. PMID: 31078628 Review. - [Crystal structures of plant uridine diphosphate-dependent glycosyltransferases].
Lü H, Xue F, Liu C, Yang M, Ma L. Lü H, et al. Sheng Wu Gong Cheng Xue Bao. 2014 Jun;30(6):838-47. Sheng Wu Gong Cheng Xue Bao. 2014. PMID: 25212002 Review. Chinese.
Cited by
- Genome-wide analysis of UDP-glycosyltransferase gene family and identification of members involved in flavonoid glucosylation in Chinese bayberry (Morella rubra).
Ren C, Cao Y, Xing M, Guo Y, Li J, Xue L, Sun C, Xu C, Chen K, Li X. Ren C, et al. Front Plant Sci. 2022 Sep 26;13:998985. doi: 10.3389/fpls.2022.998985. eCollection 2022. Front Plant Sci. 2022. PMID: 36226286 Free PMC article. - A novel glucuronosyltransferase has an unprecedented ability to catalyse continuous two-step glucuronosylation of glycyrrhetinic acid to yield glycyrrhizin.
Xu G, Cai W, Gao W, Liu C. Xu G, et al. New Phytol. 2016 Oct;212(1):123-35. doi: 10.1111/nph.14039. Epub 2016 Jun 2. New Phytol. 2016. PMID: 27252088 Free PMC article. - Structural insights into the catalytic selectivity of glycosyltransferase SgUGT94-289-3 towards mogrosides.
Cui S, Zhang S, Wang N, Su X, Luo Z, Ma X, Li M. Cui S, et al. Nat Commun. 2024 Jul 30;15(1):6423. doi: 10.1038/s41467-024-50662-w. Nat Commun. 2024. PMID: 39080270 Free PMC article. - Chemoenzymatic indican for light-driven denim dyeing.
Bidart GN, Teze D, Jansen CU, Pasutto E, Putkaradze N, Sesay AM, Fredslund F, Lo Leggio L, Ögmundarson O, Sukumara S, Qvortrup K, Welner DH. Bidart GN, et al. Nat Commun. 2024 Feb 27;15(1):1489. doi: 10.1038/s41467-024-45749-3. Nat Commun. 2024. PMID: 38413572 Free PMC article. - The crystal structure of two macrolide glycosyltransferases provides a blueprint for host cell antibiotic immunity.
Bolam DN, Roberts S, Proctor MR, Turkenburg JP, Dodson EJ, Martinez-Fleites C, Yang M, Davis BG, Davies GJ, Gilbert HJ. Bolam DN, et al. Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5336-41. doi: 10.1073/pnas.0607897104. Epub 2007 Mar 21. Proc Natl Acad Sci U S A. 2007. PMID: 17376874 Free PMC article.
References
- Achnine, L., Huhman, D.V., Farag, M.A., Sumner, L.W., Blount, J.W., and Dixon, R.A. (2005). Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J. 41, 875–887. - PubMed
- Behboudi, S., Morein, B., and Villacres-Eriksson, M.C. (1999). Quillaja saponin formulations that stimulate proinflammatory cytokines elicit a potent acquired cell-mediated immunity. Scand. J. Immunol. 50, 371–377. - PubMed
- Bowles, D., Isayenkova, J., Lim, E., and Poppenberger, B. (2005). Glycosyltransferases: Managers of small molecules. Curr. Opin. Plant Biol. 8, 254–263. - PubMed
- Breton, C., Heissigerova, H., Jeanneau, C., Moravcova, J., and Imberty, A. (2002). Comparative aspects of glycosyltransferases. Biochem. Soc. Symp. 69, 23–32. - PubMed
- Brünger, A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous