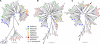Gain and loss of multiple genes during the evolution of Helicobacter pylori - PubMed (original) (raw)
Gain and loss of multiple genes during the evolution of Helicobacter pylori
Helga Gressmann et al. PLoS Genet. 2005 Oct.
Abstract
Sequence diversity and gene content distinguish most isolates of Helicobacter pylori. Even greater sequence differences differentiate distinct populations of H. pylori from different continents, but it was not clear whether these populations also differ in gene content. To address this question, we tested 56 globally representative strains of H. pylori and four strains of Helicobacter acinonychis with whole genome microarrays. Of the weighted average of 1,531 genes present in the two sequenced genomes, 25% are absent in at least one strain of H. pylori and 21% were absent or variable in H. acinonychis. We extrapolate that the core genome present in all isolates of H. pylori contains 1,111 genes. Variable genes tend to be small and possess unusual GC content; many of them have probably been imported by horizontal gene transfer. Phylogenetic trees based on the microarray data differ from those based on sequences of seven genes from the core genome. These discrepancies are due to homoplasies resulting from independent gene loss by deletion or recombination in multiple strains, which distort phylogenetic patterns. The patterns of these discrepancies versus population structure allow a reconstruction of the timing of the acquisition of variable genes within this species. Variable genes that are located within the cag pathogenicity island were apparently first acquired en bloc after speciation. In contrast, most other variable genes are of unknown function or encode restriction/modification enzymes, transposases, or outer membrane proteins. These seem to have been acquired prior to speciation of H. pylori and were subsequently lost by convergent evolution within individual strains. Thus, the use of microarrays can reveal patterns of gene gain or loss when examined within a phylogenetic context that is based on sequences of core genes.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
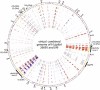
Figure 1. Genes Present and Absent in 56 Strains of H. pylori and Four Strains of H. acinonychis
CDSs used in microarrays are shown to scale along a virtual genome consisting of CDSs from both 26695 and J99 in the gene order found within 26695. Circle contents from outside to inside: (1) virtual chromosome (1.76 Mb) with ticks every 220 kb (2), GC content indicated in colors (orange, < 39%; purple, > 39%; green, rRNA genes) (3–9), numbers of missing CDSs from individual strains according to population, color-coded according to presence in both 26695 and J99 (gray) or specific to either 26695 (red) or J99 (blue). Circle, population; 3, hpAfrica2; 4, hpAfrica1; 5, hpEurope; 6, hpAsia2; 7, hpEastAsia; 8, AmerindB; 9, H. acinonychis.
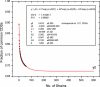
Figure 2. Extrapolated Number of Universally Present CDSs in H. pylori
The fraction of CDSs present in a sample of strains (“common CDSs”) was calculated on random samples of one to 56 strains taken without replacement. Mean fractions of common CDSs were calculated from 100 iterations of this sampling procedure. The graph shows the results of fitting an exponential decay model to these calculations, in which y0 approaches the minimum number of universally common CDSs at infinity (0.674 × 1,649 CDSs = 1,111 universally present CDSs).
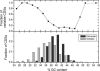
Figure 3. GC Content of CDSs That Are Universally Present or Variable within H. pylori
CDSs were binned according to GC content in steps of 2% (24–26, 26–28, etc.). Top: Fraction of all CDSs within a bin that are variable. Bottom: Fraction of universally present (n = 1,150) or variable (n = 499) CDSs by GC content. One universally present CDS with a GC content of 62% (HP0359) has been excluded from the figure.
Figure 4. Phylogenetic Structure (Neighbor-Joining Trees) According to (A) Sequences of Seven Core Genes, (B) Microarray Data Excluding cag PAI, and (C) Microarray Data Including the cag PAI for 56 Strains of H. pylori and Four Strains of H. acinonychis
Filled triangles indicate strains possessing the cag PAI, open circles indicate strains lacking it, and filled circles indicate hspAmerind strains that lack HP0536–0548 from the cag PAI. Colors indicate population assignments by Structure based on the sequence data (B. Linz, unpublished data). Numbers at the tips of the twigs are strain numbers (Table S3), while blue numbers next to nodes are bootstrap values over 75% after 250 iterations.
Similar articles
- Inter-species horizontal transfer resulting in core-genome and niche-adaptive variation within Helicobacter pylori.
Saunders NJ, Boonmee P, Peden JF, Jarvis SA. Saunders NJ, et al. BMC Genomics. 2005 Jan 27;6:9. doi: 10.1186/1471-2164-6-9. BMC Genomics. 2005. PMID: 15676066 Free PMC article. - Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island.
Devi SM, Ahmed I, Khan AA, Rahman SA, Alvi A, Sechi LA, Ahmed N. Devi SM, et al. BMC Genomics. 2006 Jul 27;7:191. doi: 10.1186/1471-2164-7-191. BMC Genomics. 2006. PMID: 16872520 Free PMC article. - A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains.
Salama N, Guillemin K, McDaniel TK, Sherlock G, Tompkins L, Falkow S. Salama N, et al. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14668-73. doi: 10.1073/pnas.97.26.14668. Proc Natl Acad Sci U S A. 2000. PMID: 11121067 Free PMC article. - Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes.
Alm RA, Trust TJ. Alm RA, et al. J Mol Med (Berl). 1999 Dec;77(12):834-46. doi: 10.1007/s001099900067. J Mol Med (Berl). 1999. PMID: 10682319 Review. - Contributions of genome sequencing to understanding the biology of Helicobacter pylori.
Ge Z, Taylor DE. Ge Z, et al. Annu Rev Microbiol. 1999;53:353-87. doi: 10.1146/annurev.micro.53.1.353. Annu Rev Microbiol. 1999. PMID: 10547695 Review.
Cited by
- Worldwide Population Structure, Long-Term Demography, and Local Adaptation of Helicobacter pylori.
Montano V, Didelot X, Foll M, Linz B, Reinhardt R, Suerbaum S, Moodley Y, Jensen JD. Montano V, et al. Genetics. 2015 Jul;200(3):947-63. doi: 10.1534/genetics.115.176404. Epub 2015 May 20. Genetics. 2015. PMID: 25995212 Free PMC article. - Prokaryotic horizontal gene transfer within the human holobiont: ecological-evolutionary inferences, implications and possibilities.
Sitaraman R. Sitaraman R. Microbiome. 2018 Sep 17;6(1):163. doi: 10.1186/s40168-018-0551-z. Microbiome. 2018. PMID: 30223892 Free PMC article. Review. - Using macro-arrays to study routes of infection of Helicobacter pylori in three families.
Raymond J, Thiberge JM, Kalach N, Bergeret M, Dupont C, Labigne A, Dauga C. Raymond J, et al. PLoS One. 2008 May 21;3(5):e2259. doi: 10.1371/journal.pone.0002259. PLoS One. 2008. PMID: 18493595 Free PMC article. - Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer.
McClain MS, Shaffer CL, Israel DA, Peek RM Jr, Cover TL. McClain MS, et al. BMC Genomics. 2009 Jan 5;10:3. doi: 10.1186/1471-2164-10-3. BMC Genomics. 2009. PMID: 19123947 Free PMC article. - A novel taxon selection method, aimed at minimizing recombination, clarifies the discovery of a new sub-population of Helicobacter pylori from Australia.
Lamichhane B, Wise MJ, Chua EG, Marshall BJ, Tay CY. Lamichhane B, et al. Evol Appl. 2019 Sep 18;13(2):278-289. doi: 10.1111/eva.12864. eCollection 2020 Feb. Evol Appl. 2019. PMID: 31993076 Free PMC article.
References
- Suerbaum S, Michetti P. Helicobacter pylori infection. New Engl J Med. 2002;347:1175–1186. - PubMed
- Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. - PubMed
- Li L, Genta RM, Go MF, Gutierrez O, Kim JG, et al. Helicobacter pylori strain and the pattern of gastritis among first-degree relatives of patients with gastric carcinoma. Helicobacter. 2002;7:349–355. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous