The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment - PubMed (original) (raw)
The DnaJ-related factor Mrj interacts with nuclear factor of activated T cells c3 and mediates transcriptional repression through class II histone deacetylase recruitment
Yan-Shan Dai et al. Mol Cell Biol. 2005 Nov.
Abstract
The calcium-regulated protein phosphatase calcineurin (PP2B) functions as a regulator of gene expression in diverse tissues through the dephosphorylation and activation of a family of transcription factors known as nuclear factor of activated T cells (NFAT). Here we show that NFATc3, in addition to being calcium responsive, is regulated through an indirect recruitment of class II histone deacetylases (HDACs). Specifically, yeast two-hybrid screening with the rel homology domain of NFATc3 identified the chaperone mammalian relative of DnaJ (Mrj) as a specific interacting factor. Mrj and NFATc3 were shown to directly associate with one another in mammalian cells and in vitro. Mrj served as a potent inhibitor of NFAT transcriptional activity within the nucleus through a mechanism involving histone deacetylase recruitment in conjunction with heat shock stimulation. Indeed, Mrj was determined to interact with class II histone deacetylases, each of which translocated to the nucleus following heat shock stimulation. Mrj also decreased NFATc3 occupancy of the tumor necrosis factor-alpha promoter in cardiomyocytes in an HDAC-dependent manner, and Mrj blocked calcineurin-induced cardiomyocyte hypertrophic growth. Conversely, small-interfering-RNA-mediated reduction of Mrj augmented NFAT transcriptional activity and spontaneously induced cardiac myocyte growth. Collectively, our results define a novel response pathway whereby NFATc3 is negatively regulated by class II histone deacetylases through the DnaJ (heat shock protein-40) superfamily member Mrj.
Figures
FIG. 1.
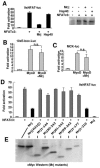
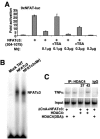
Identification of Mrj as an NFAT interacting factor. (A) A yeast two-hybrid analysis using a liquid β-galactosidase activity assay with the Mrj prey vector and full-length NFATc3 or various NFATc3 deletion mutant domains within the rel homology domain. (B) GST pull-down assay with GST or GST-Mrj and [35S]methionine-labeled NFATc3 deletion mutants. The asterisks show the position of each of the input proteins, while the arrows show the NFATc3 fragments that interact with Mrj. Ten percent of the input is shown in lanes 1 to 4. (C and D) GST pull-down assay with the indicated GST-Mrj deletion mutants in conjunction with [35S]methionine-labeled NFATc3 amino acids 405 to 700. Ten percent of the input is shown in lane 1. (E) A control showing that [35S]methionine-labeled Hsp70 interacts with the N-terminal DnaJ domain of GST-Mrj. (F) Western blot for Mrj after an in vivo immunoprecipitation assay from 10T1/2 fibroblasts that were transfected with expression vectors encoding Mrj and full-length NFATc3. (G) In vivo immunoprecipitation assay was employed in 10T1/2 cells transfected with constructs encoding four different Mrj truncation mutants and full-length NFAT. The asterisks show specific immunoprecipitation. IgG is a nonspecific control antibody. (H) Schematic representation of the different Mrj truncation mutants and their ability to interact with NFATc3.
FIG. 2.
Mrj represses NFAT-dependent transcription. (A) Relative luciferase activity in 10T1/2 cells transfected with the NFAT-dependent reporter and plasmids encoding NFATc3, Hsp40, and Mrj. The data shown were averaged from triplicate transfections (repeated twice). The panel on the right is a Western blot for NFATc3 protein from the cotransfection reactions. (B and C) Relative luciferase activity in 10T1/2 cells transfected with an E-box-dependent luciferase reporter (10xE-box-luc) or a MCK-luciferase reporter and MyoD alone or with Mrj. (D) Relative luciferase activity in 10T1/2 cells transfected with the NFAT-dependent luciferase reporter, NFATc3, and the indicated mutants of Mrj. The data shown were averaged from triplicate transfections (repeated twice). (E) Western blot (c-Myc antibody) from cells transfected with each of the Mrj mutants (all were c-Myc epitope tagged).
FIG. 3.
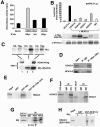
Mrj interacts with class II HDACs. (A) Deacetylase activity assay from immunoprecipitation reactions against Mrj (c-Myc antibody) or a nonspecific antibody (HA) in cardiomyocytes infected with AdMrj or a control adenovirus (AdβGal). (B) Transfection assay showing NFAT-dependent activity (9xNFAT luciferase reporter) in the presence of cotransfected NFATc3 and HDAC4/HDAC5/HDAC7/HDAC9 or Mrj. Western blotting is shown below for each transfection reaction to verify expression of HDAC4/HDAC5/HDAC7/HDAC9 (all are flag tagged) or NFATc3. (C) Coimmunoprecipitation assay followed by Western blotting for Mrj by using a c-Myc antibody (upper panel). Reactions were performed using the inputs shown (lower panels) from cells infected with AdHDAC4 alone, AdMrj alone, or both. The polyhistidine antibody served as a positive control for Mrj precipitation, the HA antibody was a negative control, and the Flag antibody showed a specific interaction between Mrj and HDAC4. (D) Coimmunoprecipitation assay followed by Western blotting for HDAC4 and NFATc3 from cotransfected Cos-7 cells with HDAC4, NFATc3, ΔCnA, and Mrj. Mrj was immunoprecipitated with a c-Myc antibody, which identified HDAC4 and NFATc3. (E) SDS-PAGE of [35S]methionine-labeled HDAC4 precipitated with GST, GST-Mrj, or GST-NFATc3 with amino acids 700 to 1075 (GST-NFΔ700-1075). Thirty percent of the input is shown. (F) Coimmunoprecipitation assay with Mrj antibody (c-Myc tag) or a nonspecific IgG, followed by Western blotting for HDAC5 or HDAC7. Cos-7 cells were first cotransfected with HDAC5 or HDAC7 in combination with Mrj. (G) Immunoprecipitation of endogenous HDAC4 (HD4), NFATc3 (NFc3), or nonspecific IgG from C2C12 protein lysates, followed by Western blotting for endogenous Mrj. (H) SDS-PAGE of [35S]methionine-labeled HDAC4 (amino acids 628 to 1084) in the presence or absence of cotranslated Mrj. These translated products were subjected to pull-down assay with GST or GST-NFATc3 (amino acids 405 to 700).
FIG. 4.
Mrj specifically interacts with HDAC4. (A) Western blot for Mrj after an in vivo immunoprecipitation assay (against HDAC4 or a nonspecific IgG) from 10T1/2 cells transfected with expression vectors encoding truncation mutants of Mrj and full-length HDAC4. (B) GST pull-down assay with the indicated Mrj mutants and [35S]methionine-labeled HDAC4 amino acids 628 to 1085. (C) A schematic representation of the different Mrj truncation mutants and their interaction with HDAC4. (D) SDS-PAGE of [35S]methionine-labeled HDAC4 truncation mutants precipitated with either GST or GST-Mrj. (E) SDS-PAGE of [35S]methionine-labeled HDAC4 deletion fragments subjected to pull-down with GST or GST-Mrj. The asterisks show the specific interactions. (F) A schematic representation of the different HDAC4 truncation mutants and their abilities to interact with Mrj.
FIG. 5.
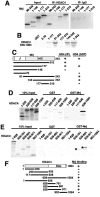
Mrj represses NFAT-dependent transcription through class II HDAC recruitment. (A) Relative luciferase activity from 10T1/2 cells transfected with the NFAT-dependent luciferase reporter, NFATc3, Mrj, and HDAC4. The asterisk signifies a P of <0.05 versus NFATc3 plus HDAC4. The lower panel shows a Western blot for NFATc3 protein from the various cotransfection conditions. (B) Relative luciferase activity from cardiomyocytes transfected with a TNF-α promoter-luciferase fusion reporter together with expression plasmids encoding NFATc3, Mrj, activated calcineurin, and HDAC4-3SA (constitutively nuclear). The asterisk signifies a P of <0.05 versus NFATc3 plus ΔCnA. The lower panel shows a Western blot for NFATc3 protein from the various cotransfection conditions. (C) SDS-PAGE of an RNase protection assay from cultured cardiomyocytes infected with the indicated adenoviruses. The position of TNF-α, GAPDH, and L32 protected fragments is shown. (D) RT-PCR for TNF-α or GAPDH mRNA levels from cardiomyocytes infected with the indicated recombinant adenoviruses. (E) ChIP assay against the TNF-α and GAPDH promoters with antibodies against Mrj, HDAC4, IgG (nonspecific), or acetyl-histone 3 (molecular weight standard is shown in the far left lane). ChIP was performed from cardiomyocytes infected with AdΔNFATc3 or AdΔCnA, in conjunction with AdMrj or AdHDAC4-3SA. (F) ChIP for the TNF-α promoter after immunoprecipitation of NFATc3 from cardiomyocytes infected with the indicated adenoviruses with or without TSA treatment.
FIG. 6.
Characterization of the NFATc3/Mrj/HDAC4 repressor mechanism. (A) Activation from a transient transfection assay in 10T1/2 cells with the NFAT-dependent luciferase reporter. TSA reversed Mrj-mediated repression of NFATc3 transactivation at 0.1 and 0.3 μg of constitutive active ΔNFATc3 (amino acids 204 to 1075) expression vector employed, although reversal was more effective at lower concentrations of Mrj. Cells were treated with or without 160 nM TSA for 16 h before harvest. (B) Electrophoretic mobility shift assay with a 32P-labeled NFAT binding site and in vitro-translated NFATc3 (amino acids 405 to 700) in the presence or absence of in vitro-translated Mrj. Mock-translated TNT lysates were used as a negative control. (C) ChIP assay to evaluate the amount of HDAC4 present at the TNF-α promoter. Cardiomyocytes were infected with AdHDAC4-3SA with or without AdNFATc3 and AdΔCnA, or infected with AdHDAC4, AdNFATc3, and AdΔCnA for 24 h and incubated continuously at 37°C or shifted to 42°C for 70 min. ChIP was performed with HDAC4 antibody.
FIG. 7.
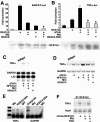
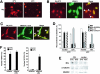
Mrj/HDAC4/NFATc3 subcellular localization. (A) Subcellular localization of Mrj in cultured cardiomyocytes at 37°C and 42°C. Myocytes were infected with an adenovirus encoding Mrj and subsequently immunostained for Mrj (red). (B) Subcellular localization of Mrj (red) and wild-type HDAC4 (green) in cardiomyocytes at 42°C (70 min of heat shock) after adenoviral overexpression of each protein together. The arrowheads show cells with colocalization, yet some cells fail to show colocalization as denoted by the asterisk. (C) Subcellular distribution of Mrj (red) and NFATc3 (green) in cardiomyocytes at 37°C previously infected with AdMrj, AdNFATc3, and AdΔCnA. (D) Graph showing quantitation of Mrj subcellular localization in either the nucleus or cytoplasm at 37°C or 42°C in cultured cardiomyocytes infected with the indicated combination of adenoviruses. (E) Western blot for Mrj, α-tubulin, and HDAC2 from cardiomyocytes infected with AdMrj. Nuclear and cytoplasmic extracts were generated from cells incubated at 37°C or 42°C (40 min of heat shock). (F) Graph showing percentages of cardiomyocytes with nuclear HDAC4 (left) or NFATc3 (right) at 37°C and 42°C after adenoviral infection to overexpress HDAC4 alone, HDAC4 with Mrj, or NFATc3 with or without Mrj. Subcellular localizations were quantified in 200 cells each in three independent experiments.
FIG. 8.
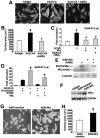
Mrj represses calcineurin-induced cardiomyocyte hypertrophy. (A) Pictures of α-actinin-stained neonatal cardiomyocytes infected with the indicated recombinant adenoviruses. (B) Quantification of cardiomyocyte size. *, P < 0.05 versus Adβgal; #, P < 0.05 versus AdΔCnA. (C) Relative luciferase activity from cardiomyocytes transfected with the NFAT-dependent luciferase reporter plasmid along with expression plasmids encoding activated calcineurin, Mrj, and HDAC4-3SA. *, P < 0.05 versus reporter only; #, P < 0.05 versus ΔCnA. (D) Relative luciferase activity from 10T1/2 cells transfected with the NFAT-dependent luciferase reporter plasmid and expression plasmids encoding NFATc3, Mrj, siMrj (SiMrJ), or a random siRNA (Si-random) sequence. *, P < 0.05 versus NFATc3 with random siRNA sequence. (E) Western blot analysis of Mrj protein from adenoviral infected neonatal cardiomyocytes expressing siRNA-Mrj or Mrj. Endo, endogenous Mrj protein. The adenoviral-encoded Mrj is larger due to epitope tags. α-Tubulin, as a loading control, is shown. (F) RT-PCR for TNF-α or GAPDH mRNA levels from cardiomyocytes infected with Adβgal (lane 1), AdΔCnA, AdNFATc3, and AdSi-random (lane 2), and AdΔCnA, AdNFATc3, and AdSi-Mrj (lane 3). (G) Pictures of α-actinin-stained neonatal cardiomyocytes infected with the indicated recombinant adenoviruses. (H) Quantification of cardiomyocyte size. *, P < 0.05 versus AdSi-random.
FIG. 9.
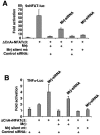
(A) Activation (_n_-fold) from a transient transfection assay in 10T1/2 cells with the NFAT-dependent luciferase reporter. The experiment shows the specificity of Mrj-siRNA by cotransfecting a vector that encodes wild-type Mrj with silent mutations in the area recognized by the Mrj-siRNA. Mrj-siRNA partially reverses Mrj-dependent inhibition of NFAT activity, but this partial reversal was not observed with cotransfected silent mutant Mrj. Luciferase activity was determined 60 h after transfection. (B) Relative luciferase activity in 10T1/2 cells transfected with a TNF-α-luciferase reporter in conjunction with NFATc3 and ΔCnA in the presence of a negative control siRNA, Mrj-siRNA, or the silent mutant Mrj as in panel A. Mrj-siRNA up-regulated TNF-α reporter activity.
Similar articles
- Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth.
Sanna B, Bueno OF, Dai YS, Wilkins BJ, Molkentin JD. Sanna B, et al. Mol Cell Biol. 2005 Feb;25(3):865-78. doi: 10.1128/MCB.25.3.865-878.2005. Mol Cell Biol. 2005. PMID: 15657416 Free PMC article. - Calcineurin regulates NFAT-dependent iNOS expression and protection of cardiomyocytes: co-operation with Src tyrosine kinase.
Obasanjo-Blackshire K, Mesquita R, Jabr RI, Molkentin JD, Hart SL, Marber MS, Xia Y, Heads RJ. Obasanjo-Blackshire K, et al. Cardiovasc Res. 2006 Sep 1;71(4):672-83. doi: 10.1016/j.cardiores.2006.05.026. Epub 2006 Jun 3. Cardiovasc Res. 2006. PMID: 16828070 - HTRP--an immediate-early gene product induced by HSV1 infection in human embryo fibroblasts, is involved in cellular co-repressors.
Li JF, Liu LD, Ma SH, Che YC, Wang LC, Dong CH, Zhao HL, Liao Y, Li QH. Li JF, et al. J Biochem. 2004 Aug;136(2):169-76. doi: 10.1093/jb/mvh108. J Biochem. 2004. PMID: 15496587 - Calcium-calcineurin signaling in the regulation of cardiac hypertrophy.
Wilkins BJ, Molkentin JD. Wilkins BJ, et al. Biochem Biophys Res Commun. 2004 Oct 1;322(4):1178-91. doi: 10.1016/j.bbrc.2004.07.121. Biochem Biophys Res Commun. 2004. PMID: 15336966 Review. - Transcription regulation by histone deacetylases.
Wang S, Yan-Neale Y, Zeremski M, Cohen D. Wang S, et al. Novartis Found Symp. 2004;259:238-45; discussion 245-8, 285-8. Novartis Found Symp. 2004. PMID: 15171258 Review.
Cited by
- The role of redox modulation of class II histone deacetylases in mediating pathological cardiac hypertrophy.
Oka S, Ago T, Kitazono T, Zablocki D, Sadoshima J. Oka S, et al. J Mol Med (Berl). 2009 Aug;87(8):785-91. doi: 10.1007/s00109-009-0471-2. Epub 2009 May 8. J Mol Med (Berl). 2009. PMID: 19424677 Review. - DNAJB6 induces degradation of beta-catenin and causes partial reversal of mesenchymal phenotype.
Mitra A, Menezes ME, Shevde LA, Samant RS. Mitra A, et al. J Biol Chem. 2010 Aug 6;285(32):24686-94. doi: 10.1074/jbc.M109.094847. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522561 Free PMC article. - Large isoform of MRJ (DNAJB6) reduces malignant activity of breast cancer.
Mitra A, Fillmore RA, Metge BJ, Rajesh M, Xi Y, King J, Ju J, Pannell L, Shevde LA, Samant RS. Mitra A, et al. Breast Cancer Res. 2008;10(2):R22. doi: 10.1186/bcr1874. Epub 2008 Mar 7. Breast Cancer Res. 2008. PMID: 18328103 Free PMC article. - Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy.
Sarparanta J, Jonson PH, Golzio C, Sandell S, Luque H, Screen M, McDonald K, Stajich JM, Mahjneh I, Vihola A, Raheem O, Penttilä S, Lehtinen S, Huovinen S, Palmio J, Tasca G, Ricci E, Hackman P, Hauser M, Katsanis N, Udd B. Sarparanta J, et al. Nat Genet. 2012 Feb 26;44(4):450-5, S1-2. doi: 10.1038/ng.1103. Nat Genet. 2012. PMID: 22366786 Free PMC article. - SPARC promotes insulin secretion through down-regulation of RGS4 protein in pancreatic β cells.
Hu L, He F, Huang M, Zhao Q, Cheng L, Said N, Zhou Z, Liu F, Dai YS. Hu L, et al. Sci Rep. 2020 Oct 16;10(1):17581. doi: 10.1038/s41598-020-74593-w. Sci Rep. 2020. PMID: 33067534 Free PMC article.
References
- Avots, A., M. Buttmann, S. Chuvpilo, C. Escher, U. Smola, A. J. Bannister, U. R. Rapp, T. Kouzarides, and E. Serfling. 1999. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity 10:515-524. - PubMed
- Baksh, S., H. R. Widlund, A. A. Frazer-Abel, J. Du, S. Fosmire, D. E. Fisher, J. A. DeCaprio, J. F. Modiano, and S. J. Burakoff. 2002. NFATc2-mediated repression of cyclin-dependent kinase 4 expression. Mol. Cell 10:1071-1081. - PubMed
- Chuang, J. Z., H. Zhou, M. Zhu, S. H. Li, X. J. Li, and C. H. Sung. 2002. Characterization of a brain-enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J. Biol. Chem. 277:19831-19838. - PubMed
- Crabtree, G. R. 1999. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96:611-614. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous