An analysis of the feasibility of short read sequencing - PubMed (original) (raw)
An analysis of the feasibility of short read sequencing
Nava Whiteford et al. Nucleic Acids Res. 2005.
Abstract
Several methods for ultra high-throughput DNA sequencing are currently under investigation. Many of these methods yield very short blocks of sequence information (reads). Here we report on an analysis showing the level of genome sequencing possible as a function of read length. It is shown that re-sequencing and de novo sequencing of the majority of a bacterial genome is possible with read lengths of 20-30 nt, and that reads of 50 nt can provide reconstructed contigs (a contiguous fragment of sequence data) of 1000 nt and greater that cover 80% of human chromosome 1.
Figures
Figure 1
Percentage of unique reads as a function of read length for (a) λ-phage and (b) E.coli K12. The dashed curves show results for randomly generated sequences of the same size, which are content-biased to yield the same relative proportion of nucleotides given in ref. (27).
Figure 2
(a) Percentage of unique sub-sequences (U) for varying read length (l), the solid line shows uniqueness in the whole human genome, the dashed line shows uniqueness in human chromosome 1. (b) Percentage of human chromosome 1 covered by contigs greater than a threshold length as a function of read length. The horizontal axis starts at 18 nt, due to the limitations of reassembly below this length.
Figure 3
Percentage of the E.coli genome covered by contigs greater than a threshold length as a function of read length.
Figure 4
(a) Percentage of unique sub-sequences (U) for varying read length (l), in the C.elegans genome. (b) Percentage of the C.elegans genome covered by contigs greater than a threshold length as a function of read length. The horizontal axis starts at 18 nt, due to the limitations of reassembly below this length.
Figure 5
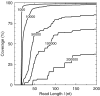
Reassembled contigs longer than 200 nt in the 81 090 nt of the BRCA1 gene. Reassembly was simulated from 25, 50 and 100 nt reads covering the whole of chromosome 17. Reassembled contigs are shown in alternating black and grey. Contigs maybe next to each other, or overlapping slightly without an unambiguous overlap existing between the contigs.
Similar articles
- Whole genome sequencing.
Ng PC, Kirkness EF. Ng PC, et al. Methods Mol Biol. 2010;628:215-26. doi: 10.1007/978-1-60327-367-1_12. Methods Mol Biol. 2010. PMID: 20238084 Review. - Structural variation analysis with strobe reads.
Ritz A, Bashir A, Raphael BJ. Ritz A, et al. Bioinformatics. 2010 May 15;26(10):1291-8. doi: 10.1093/bioinformatics/btq153. Epub 2010 Apr 8. Bioinformatics. 2010. PMID: 20378554 - Reptile: representative tiling for short read error correction.
Yang X, Dorman KS, Aluru S. Yang X, et al. Bioinformatics. 2010 Oct 15;26(20):2526-33. doi: 10.1093/bioinformatics/btq468. Epub 2010 Aug 16. Bioinformatics. 2010. PMID: 20834037 - Rapid hybrid de novo assembly of a microbial genome using only short reads: Corynebacterium pseudotuberculosis I19 as a case study.
Cerdeira LT, Carneiro AR, Ramos RT, de Almeida SS, D'Afonseca V, Schneider MP, Baumbach J, Tauch A, McCulloch JA, Azevedo VA, Silva A. Cerdeira LT, et al. J Microbiol Methods. 2011 Aug;86(2):218-23. doi: 10.1016/j.mimet.2011.05.008. Epub 2011 May 18. J Microbiol Methods. 2011. PMID: 21620904 - Whole-genome re-sequencing.
Bentley DR. Bentley DR. Curr Opin Genet Dev. 2006 Dec;16(6):545-52. doi: 10.1016/j.gde.2006.10.009. Epub 2006 Oct 18. Curr Opin Genet Dev. 2006. PMID: 17055251 Review.
Cited by
- Origin of multiple periodicities in the Fourier power spectra of the Plasmodium falciparum genome.
Nunes MC, Wanner EF, Weber G. Nunes MC, et al. BMC Genomics. 2011 Dec 22;12 Suppl 4(Suppl 4):S4. doi: 10.1186/1471-2164-12-S4-S4. Epub 2011 Dec 22. BMC Genomics. 2011. PMID: 22369134 Free PMC article. - The theory of discovering rare variants via DNA sequencing.
Wendl MC, Wilson RK. Wendl MC, et al. BMC Genomics. 2009 Oct 20;10:485. doi: 10.1186/1471-2164-10-485. BMC Genomics. 2009. PMID: 19843339 Free PMC article. - Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler.
Zerbino DR, McEwen GK, Margulies EH, Birney E. Zerbino DR, et al. PLoS One. 2009 Dec 22;4(12):e8407. doi: 10.1371/journal.pone.0008407. PLoS One. 2009. PMID: 20027311 Free PMC article. - Computational methods for detecting copy number variations in cancer genome using next generation sequencing: principles and challenges.
Liu B, Morrison CD, Johnson CS, Trump DL, Qin M, Conroy JC, Wang J, Liu S. Liu B, et al. Oncotarget. 2013 Nov;4(11):1868-81. doi: 10.18632/oncotarget.1537. Oncotarget. 2013. PMID: 24240121 Free PMC article. Review. - The long march: a sample preparation technique that enhances contig length and coverage by high-throughput short-read sequencing.
Sorber K, Chiu C, Webster D, Dimon M, Ruby JG, Hekele A, DeRisi JL. Sorber K, et al. PLoS One. 2008;3(10):e3495. doi: 10.1371/journal.pone.0003495. Epub 2008 Oct 22. PLoS One. 2008. PMID: 18941527 Free PMC article.
References
- Shendure J., Mitra R.D., Church G.M. Advanced sequencing technologies: methods and goals. Nature Rev. Gen. 2004;5:335–344. - PubMed
- Brenner S., Johnson M., Bridgham J., Golda G., Lloyd D., Johnson D., Luo S., McCurdy S., Foy M., Ewan M., et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2003;18:630–634. - PubMed
- Kling J. Ultrafast DNA sequencing. Nat. Biotechol. 2003;21:1425–1427. - PubMed
- Ronaghi M., Uhlen M., Nyren P. DNA sequencing: a sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials