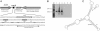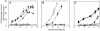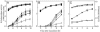Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0 - PubMed (original) (raw)
Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0
Elisabeth Kay et al. Proc Natl Acad Sci U S A. 2005.
Abstract
In many Gram-negative bacteria, the GacS/GacA two-component system positively controls the expression of extracellular products or storage compounds. In the plant-beneficial rhizosphere bacterium Pseudomonas fluorescens CHA0, the GacS/GacA system is essential for the production of antibiotic compounds and hence for biological control of root-pathogenic fungi. The small (119-nt) RNA RsmX discovered in this study, together with RsmY and RsmZ, forms a triad of GacA-dependent small RNAs, which sequester the RNA-binding proteins RsmA and RsmE and thereby antagonize translational repression exerted by these proteins in strain CHA0. This small RNA triad was found to be both necessary and sufficient for posttranscriptional derepression of biocontrol factors and for protection of cucumber from Pythium ultimum. The same three small RNAs also positively regulated swarming motility and the synthesis of a quorum-sensing signal, which is unrelated to N-acyl-homoserine lactones, and which autoinduces the Gac/Rsm cascade. Expression of RsmX and RsmY increased in parallel throughout cell growth, whereas RsmZ was produced during the late growth phase. This differential expression is assumed to facilitate fine tuning of GacS/A-controlled cell population density-dependent regulation in P. fluorescens.
Figures
Fig. 1.
The rsmX gene is not linked to gacS and gacA, depends on gacA function for expression, and shows characteristic unpaired GGA motifs. (A) Organization of the rsmX region of P. fluorescens CHA0. The -35 and -10 promoter sites are indicated with gray boxes. The palindromic sequence boxed from -70 to -53 denotes the upstream activating sequence (UAS), the putative GacA regulatory site. A sequence of 311 nucleotides (rhomboid box), which is absent from strain Pf-5, does not present any homology with a known sequence. (B) Northern blot showing the differential temporal accumulation of RsmX. Total RNA was extracted from CHA0 (wild type; lanes 1–3), CHA89 (gacA; lanes 4–6). OD600 values at the time of harvesting were, in lanes 1 and 4, 0.5; in lanes 2 and 5, 1.9; and in lanes 3 and 6, 2.5. (C) Predicted secondary structure of RsmX at 30°C using the
mfold
program (
www.bioinfo.rpi.edu/applications/mfold
).
Fig. 2.
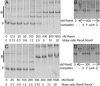
RsmA and RsmE bind to RsmX, as demonstrated by RNA gel mobility-shift analysis. RsmX was synthesized in vitro by T7 RNA polymerase in the presence of [α-33P]-UTP. Labeled RsmX (80 nM) was incubated with the concentrations of RsmA or RsmE indicated in A and C, respectively. The positions of free (F) and bound (B) RNA species are indicated. Unlabeled competitor RNAs (X = RsmX, Y = RsmY, and Z = RsmZ and CarA leader) were synthesized following the same protocol but with unlabeled UTP. Labeled RsmX (80 nM) with either specific (RsmX, RsmY, or RsmZ) or nonspecific (CarA leader) non-labeled competitors (300 nM) was incubated with 800 nM RsmA and 300 nM RsmE in B and D, respectively.
Fig. 3.
The expression of the rsmX gene is controlled by GacA, RsmA, RsmE, and the CHA0 signal in P. fluorescens CHA0. (A) Activation of rsmX expression by RsmA and RsmE. β-Galactosidase activities of a transcriptional rsmX-lacZ fusion carried by pME7317 were determined in the wild type (CHA0; squares), a gacA mutant (CHA89; open diamonds), an rsmE mutant (CHA1003; diamonds), an rsmA mutant (CHA1076; triangles), an rsmAE double mutant (CHA1009; open circles), and a gacS rsmAE mutant (CHA1008; open triangles). (B) Activation of rsmX expression by the signal from CHA0. β-Galactosidase activities of an rsmX-lacZ fusion were determined in the wild-type strain without (squares) or with extract (open squares) and the gacA mutant without (triangles) or with extract (open triangles). (C) Differential temporal expression of rsmX, rsmY, and rsmZ. β-Galactosidase activities of the transcriptional rsmX-lacZ (pME7317; squares), rsmY-lacZ [pME6916 (18), diamonds], and rsmZ-lacZ [pME6091 (14), triangles] were determined in strain CHA0 (wild-type; filled symbols) and in the gacA mutant CHA89 (empty symbols).
Fig. 4.
Deletion of rsmX, rsmY, and rsmZ results in low expression of target genes similar to that found in a gacA mutant. (A) Expression of a chromosomal hcnA_′-′_lacZ translational fusion and growth of the wild type (CHA207; squares), a gacA mutant (CHA89.207; open diamonds), an rsmX mutant (CHA1142; diamonds), an rsmY mutant (CHA823; circles), an rsmZ mutant (CHA811; triangles), an rsmY rsmZ mutant (CHA826; open squares), and an rsmX rsmY rsmZ mutant (CHA1145; open triangles). (B) Expression of a chromosomal aprA_′-′_lacZ translational fusion and growth in the wild-type (CHA805; squares), a gacS mutant (CHA806; open diamonds), an rsmX mutant (CHA1143; diamonds), an rsmY mutant (CHA824; circles), an rsmZ mutant (CHA812; triangles), an rsmY rsmZ mutant (CHA827; open squares), and an rsmX rsmY rsmZ mutant (CHA1146; open triangles). (C) Expression of a chromosomal rsmE_′-′_lacZ fusion in the wild type (CHA1134; squares), a gacA mutant (CHA1136; diamonds), and an rsmX rsmY rsmZ mutant (CHA1164; triangles). Growth is indicated by dotted lines.
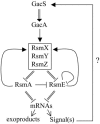
Fig. 5.
Model for gene regulation in the Gac/Rsm system of P. fluorescens. —, direct or indirect regulation; =, physical interaction; →, positive effect; ⊣, negative effect. A physical interaction of the signal with GacS has not been demonstrated experimentally.
Similar articles
- RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0.
Valverde C, Heeb S, Keel C, Haas D. Valverde C, et al. Mol Microbiol. 2003 Nov;50(4):1361-79. doi: 10.1046/j.1365-2958.2003.03774.x. Mol Microbiol. 2003. PMID: 14622422 - Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0.
Heeb S, Blumer C, Haas D. Heeb S, et al. J Bacteriol. 2002 Feb;184(4):1046-56. doi: 10.1128/jb.184.4.1046-1056.2002. J Bacteriol. 2002. PMID: 11807065 Free PMC article. - Posttranscriptional repression of GacS/GacA-controlled genes by the RNA-binding protein RsmE acting together with RsmA in the biocontrol strain Pseudomonas fluorescens CHA0.
Reimmann C, Valverde C, Kay E, Haas D. Reimmann C, et al. J Bacteriol. 2005 Jan;187(1):276-85. doi: 10.1128/JB.187.1.276-285.2005. J Bacteriol. 2005. PMID: 15601712 Free PMC article. - Signal transduction in plant-beneficial rhizobacteria with biocontrol properties.
Haas D, Keel C, Reimmann C. Haas D, et al. Antonie Van Leeuwenhoek. 2002 Aug;81(1-4):385-95. doi: 10.1023/a:1020549019981. Antonie Van Leeuwenhoek. 2002. PMID: 12448737 Review. - Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour.
Lapouge K, Schubert M, Allain FH, Haas D. Lapouge K, et al. Mol Microbiol. 2008 Jan;67(2):241-53. doi: 10.1111/j.1365-2958.2007.06042.x. Epub 2007 Nov 28. Mol Microbiol. 2008. PMID: 18047567 Review.
Cited by
- A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence.
Balasubramanian D, Schneper L, Kumari H, Mathee K. Balasubramanian D, et al. Nucleic Acids Res. 2013 Jan 7;41(1):1-20. doi: 10.1093/nar/gks1039. Epub 2012 Nov 11. Nucleic Acids Res. 2013. PMID: 23143271 Free PMC article. Review. - The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins.
Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors O. Jonas K, et al. Mol Microbiol. 2008 Oct;70(1):236-57. doi: 10.1111/j.1365-2958.2008.06411.x. Epub 2008 Aug 18. Mol Microbiol. 2008. PMID: 18713317 Free PMC article. - Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation.
Urban JH, Vogel J. Urban JH, et al. PLoS Biol. 2008 Mar 18;6(3):e64. doi: 10.1371/journal.pbio.0060064. PLoS Biol. 2008. PMID: 18351803 Free PMC article. - CrcZ and CrcX regulate carbon source utilization in Pseudomonas syringae pathovar tomato strain DC3000.
Filiatrault MJ, Stodghill PV, Wilson J, Butcher BG, Chen H, Myers CR, Cartinhour SW. Filiatrault MJ, et al. RNA Biol. 2013 Feb;10(2):245-55. doi: 10.4161/rna.23019. Epub 2013 Jan 25. RNA Biol. 2013. PMID: 23353577 Free PMC article. - Small RNA-dependent expression of secondary metabolism is controlled by Krebs cycle function in Pseudomonas fluorescens.
Takeuchi K, Kiefer P, Reimmann C, Keel C, Dubuis C, Rolli J, Vorholt JA, Haas D. Takeuchi K, et al. J Biol Chem. 2009 Dec 11;284(50):34976-85. doi: 10.1074/jbc.M109.052571. Epub 2009 Oct 19. J Biol Chem. 2009. PMID: 19840935 Free PMC article.
References
- Gottesman, S. (2004) Annu. Rev. Microbiol. 58, 303-328. - PubMed
- Storz, G., Opdyke, J. A. & Zhang, A. (2004) Curr. Opin. Microbiol. 7, 140-144. - PubMed
- Majdalani, N., Vanderpool, C. & Gottesman, S. (2005) Crit. Rev. Biochem. Mol. Biol. 40, 93-113. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials