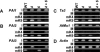H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases - PubMed (original) (raw)
H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases
Michelle L Ebbs et al. Mol Cell Biol. 2005 Dec.
Abstract
Transcribed inverted repeats are potent triggers for RNA interference and RNA-directed DNA methylation in plants through the production of double-stranded RNA (dsRNA). For example, a transcribed inverted repeat of endogenous genes in Arabidopsis thaliana, PAI1-PAI4, guides methylation of itself as well as two unlinked duplicated PAI genes, PAI2 and PAI3. In previous work, we found that mutations in the SUVH4/KYP histone H3 lysine 9 (H3 K9) methyltransferase cause a loss of DNA methylation on PAI2 and PAI3, but not on the inverted repeat. Here we use chromatin immunoprecipitation analysis to show that the transcribed inverted repeat carries H3 K9 methylation, which is maintained even in an suvh4 mutant. PAI1-PAI4 H3 K9 methylation and DNA methylation are also maintained in an suvh6 mutant, which is defective for a gene closely related to SUVH4. However, both epigenetic modifications are reduced at this locus in an suvh4 suvh6 double mutant. In contrast, SUVH6 does not play a significant role in maintenance of H3 K9 or DNA methylation on PAI2, transposon sequences, or centromere repeat sequences. Thus, SUVH6 is preferentially active at a dsRNA source locus versus targets for RNA-directed chromatin modifications.
Figures
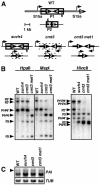
FIG. 1.
PAI gene DNA methylation patterning and transcriptional activity in DNA methylation-deficient mutants. (A) Diagram summary of DNA methylation patterns and transcriptional activity for PAI1-PAI4 and PAI2. Each PAI sequence (labeled 1, 2, or 4), including regions of identity upstream and downstream of the coding region, or the S15a gene (labeled S) is indicated by a thick black arrow. The 2.9-kb unmethylated direct repeat regions flanking PAI1-PAI4 are indicated by open bars. Regions of DNA methylation are shown boxed by a solid line for CG and non-CG methylation or by a dashed line for CG methylation. An arrow indicates transcription, and an X indicates silencing. Sites of PCR amplification used for ChIP analysis are indicated with gray bars on the diagram for wild-type WS (WT), with P1 indicating the PAI1 site, P2 indicating the PAI2 site, and S15a indicating the S15a transcription start sites. (B) Southern blot assays for PAI DNA methylation patterning. Genomic DNA from the indicated strains was cleaved with HpaII, MspI, or HincII and used in Southern blot analysis with a PAI1 cDNA probe. P1-P4 indicates PAI1-PAI4, P2 indicates PAI2, and P3 indicates PAI3, with bands diagnostic of methylation on _PAI_-internal sites denoted with asterisks. (C) PAI1 transcript levels are not significantly altered by changes in internal DNA methylation patterns at the PAI1-PAI4 transcribed inverted repeat. Total RNA was isolated from 3-week-old plants of the indicated strains and used to prepare duplicate gel blots that were probed with a PAI1 cDNA probe (PAI) or a β-tubulin (TUB) probe as a loading control. Four strains were used; WT is wild type WS, suvh4 is WS suvh4R302* (25), cmt3 is WS cmt3i11a (2), and cmt3 met1 is _pai1 cmt3i11a met1_-1 (Materials and Methods). The arrowhead in the left margin indicates the position of a 1,900-nt 5′ splice variant unique to PAI1 (28). Note that in the wild-type WS the major PAI transcript pool consists of two other PAI1 5′ splice variants that are both approximately 1,200 nt. In the three mutant backgrounds where additional PAI genes are demethylated, PAI transcripts initiating in proximal PAI promoter sequences also give rise to approximately 1,200-nt transcripts that contribute to the intensity of the smaller-molecular-size pool.
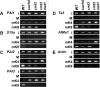
FIG. 2.
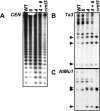
PAI gene H3 mK4 and H3 mK9 patterning in DNA methylation-deficient mutants. Primer sets specific for (A) the PAI-PAI4 transcribed inverted repeat, (B) the unmethylated S15a transcription start regions flanking the PAI1-PAI4 inverted repeat, (C) the PAI2 and PAI3 targets for PAI1-PAI4 RNA-directed DNA methylation, (D) the Ta3 retrotransposon and AtMu1 DNA transposon, and (E) the unmethylated transcribed Actin gene were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (mK4), or chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (mK9) from the indicated strains. Four strains were used: WT is wild-type WS, suvh4 is WS suvh4R302* (25), cmt3 is WS cmt3i11a (2), and cmt3 met1 is _pai1 cmt3i11a met1_-1 (Materials and Methods). GelStar-stained PCR products are shown. These results were reproduced in three independent experiments, with a representative data set shown.
FIG. 3.
PAI gene H3 mK4 and H3 mK9 patterning in suvh6 and suvh4 suvh6 mutants. Primer sets specific for (A) the PAI-PAI4 transcribed inverted repeat, (B) the PAI2 and PAI3 targets for PAI1-PAI4 RNA-directed DNA methylation, (C) the Ta3 retrotransposon and AtMu1 DNA transposon, and (D) the unmethylated transcribed Actin gene were used to amplify PCR products from total input chromatin (I), no-antibody mock precipitation control (M), chromatin immunoprecipitated with H3 anti-dimethyl K4 antibodies (mK4), or chromatin immunoprecipitated with H3 anti-dimethyl K9 antibodies (mK9) from the indicated strains. Four strains, wild-type WS (WT) and three mutants, were used. All mutations assayed were in the WS pai1 background, with 4 indicating suvh4, 6 indicating suvh6, and 4 + 6 indicating suvh4 suvh6. GelStar-stained PCR products are shown. These results were reproduced in three independent experiments, with a representative data set shown.
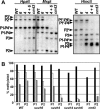
FIG. 4.
The suvh4 suvh6 double mutant displays loss of DNA methylation at the PAI1-PAI4 inverted repeat. (A) Southern blot assays for PAI DNA methylation patterning in suvh6 and suvh4 suvh6 mutants. Genomic DNA from the indicated strains was cleaved with HpaII, MspI, or HincII and used in Southern blot analysis with a PAI1 cDNA probe. P1-P4 indicates PAI1-PAI4, P2 indicates PAI2, and P3 indicates PAI3, with bands diagnostic of methylation on _PAI_-internal sites denoted with asterisks. All mutations assayed were in the WS pai1 background, with WT indicating wild type for SUVH4 and SUVH6, 4 indicating suvh4, 6 indicating suvh6, and 4 + 6 indicating suvh4 suvh6. (B) Bisulfite genomic sequencing of DNA methylation patterning on the PAI1 and PAI2 proximal promoters in suvh6 and suvh4 suvh6 mutants. Eight independent top-strand clones were sequenced for PAI1 (P1) or PAI2 (P2) from the same DNA samples used in panel A. The percentage of 5-methyl-cytosines out of total cytosines sequenced within the region of PAI sequence identity (344 bp for PAI1 or 338 bp for PAI2) is shown, divided into the following contexts: CG (black), CNG (white), or other (gray). Data for wild-type WS (WT) and cmt3 and suvh4 mutants are from previous publications (2, 24, 25).
FIG. 5.
The suvh6 mutation does not suppress PAI2 silencing. Representative 2-week-old seedlings of the indicated genotypes are shown photographed under visible light (top row) or short-wave UV light (bottom row). WT is wild-type WS.
FIG. 6.
Centromere and transposon non-CG methylation patterns in suvh6 and suvh4 suvh6 mutants. DNA from the indicated strains was cleaved with MspI (A and B) or HindIII plus MspI (C) and used in Southern blot analysis with a 180-bp centromere repeat sequence probe (CEN) (A), a Ta3 probe (B), or an AtMu1 probe (C). Arrowheads in the left margin indicate the positions of fully cleaved bands. All mutations assayed were in the WS pai1 background, with WT indicating wild type for SUVH4 and SUVH6, 4 indicating suvh4, 6 indicating suvh6, and 4 + 6 indicating suvh4 suvh6.
Similar articles
- Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase.
Ebbs ML, Bender J. Ebbs ML, et al. Plant Cell. 2006 May;18(5):1166-76. doi: 10.1105/tpc.106.041400. Epub 2006 Mar 31. Plant Cell. 2006. PMID: 16582009 Free PMC article. - Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase.
Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Jackson JP, et al. Nature. 2002 Apr 4;416(6880):556-60. doi: 10.1038/nature731. Epub 2002 Mar 17. Nature. 2002. PMID: 11898023 - Small RNAs prevent transcription-coupled loss of histone H3 lysine 9 methylation in Arabidopsis thaliana.
Enke RA, Dong Z, Bender J. Enke RA, et al. PLoS Genet. 2011 Oct;7(10):e1002350. doi: 10.1371/journal.pgen.1002350. Epub 2011 Oct 27. PLoS Genet. 2011. PMID: 22046144 Free PMC article. - DNA and histone methylation in plants.
Tariq M, Paszkowski J. Tariq M, et al. Trends Genet. 2004 Jun;20(6):244-51. doi: 10.1016/j.tig.2004.04.005. Trends Genet. 2004. PMID: 15145577 Review. - The many faces of histone lysine methylation.
Lachner M, Jenuwein T. Lachner M, et al. Curr Opin Cell Biol. 2002 Jun;14(3):286-98. doi: 10.1016/s0955-0674(02)00335-6. Curr Opin Cell Biol. 2002. PMID: 12067650 Review.
Cited by
- DNA polymerase epsilon binds histone H3.1-H4 and recruits MORC1 to mediate meiotic heterochromatin condensation.
Wang C, Huang J, Li Y, Zhang J, He C, Li T, Jiang D, Dong A, Ma H, Copenhaver GP, Wang Y. Wang C, et al. Proc Natl Acad Sci U S A. 2022 Oct 25;119(43):e2213540119. doi: 10.1073/pnas.2213540119. Epub 2022 Oct 19. Proc Natl Acad Sci U S A. 2022. PMID: 36260743 Free PMC article. - Epigenetic and epigenomic variation in Arabidopsis thaliana.
Schmitz RJ, Ecker JR. Schmitz RJ, et al. Trends Plant Sci. 2012 Mar;17(3):149-54. doi: 10.1016/j.tplants.2012.01.001. Epub 2012 Feb 17. Trends Plant Sci. 2012. PMID: 22342533 Free PMC article. Review. - Reprogramming of Histone H3 Lysine Methylation During Plant Sexual Reproduction.
Fang H, Shao Y, Wu G. Fang H, et al. Front Plant Sci. 2021 Nov 30;12:782450. doi: 10.3389/fpls.2021.782450. eCollection 2021. Front Plant Sci. 2021. PMID: 34917115 Free PMC article. Review. - DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns.
Rigal M, Kevei Z, Pélissier T, Mathieu O. Rigal M, et al. EMBO J. 2012 Jun 29;31(13):2981-93. doi: 10.1038/emboj.2012.141. Epub 2012 May 11. EMBO J. 2012. PMID: 22580822 Free PMC article. - Molecular mechanisms of epigenetic variation in plants.
Fujimoto R, Sasaki T, Ishikawa R, Osabe K, Kawanabe T, Dennis ES. Fujimoto R, et al. Int J Mol Sci. 2012;13(8):9900-9922. doi: 10.3390/ijms13089900. Epub 2012 Aug 8. Int J Mol Sci. 2012. PMID: 22949838 Free PMC article. Review.
References
- Baumbusch, L. O., T. Thorstensen, V. Krauss, A. Fischer, K. Naumann, R. Assalkhou, I. Schulz, G. Reuter, and R. B. Aalen. 2001. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29:4319-4333. - PMC - PubMed
- Bender, J., and G. R. Fink. 1995. Epigenetic control of an endogenous gene family is revealed by a novel blue fluorescent mutant of Arabidopsis. Cell 83:725-734. - PubMed
- Cao, X., W. Aufsatz, D. Zilberman, M. F. Mette, M. S. Huang, M. Matzke, and S. E. Jacobsen. 2003. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13:2212-2217. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM61148/GM/NIGMS NIH HHS/United States
- T32 CA009110/CA/NCI NIH HHS/United States
- R01 GM061148/GM/NIGMS NIH HHS/United States
- T32 CA09110/CA/NCI NIH HHS/United States
- T32 ES007141/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous